Viral and Host Transcriptomes in SARS-CoV-2-Infected Human Lung Cells
Xuefeng Wang, Yudong Zhao, Feihu Yan, Tiecheng Wang, Weiyang Sun, Na Feng, Wenqi Wang, Hongmei Wang, Hongbin He, Songtao Yang, Xianzhu Xia, Yuwei Gao
doi:10.1128/JVI
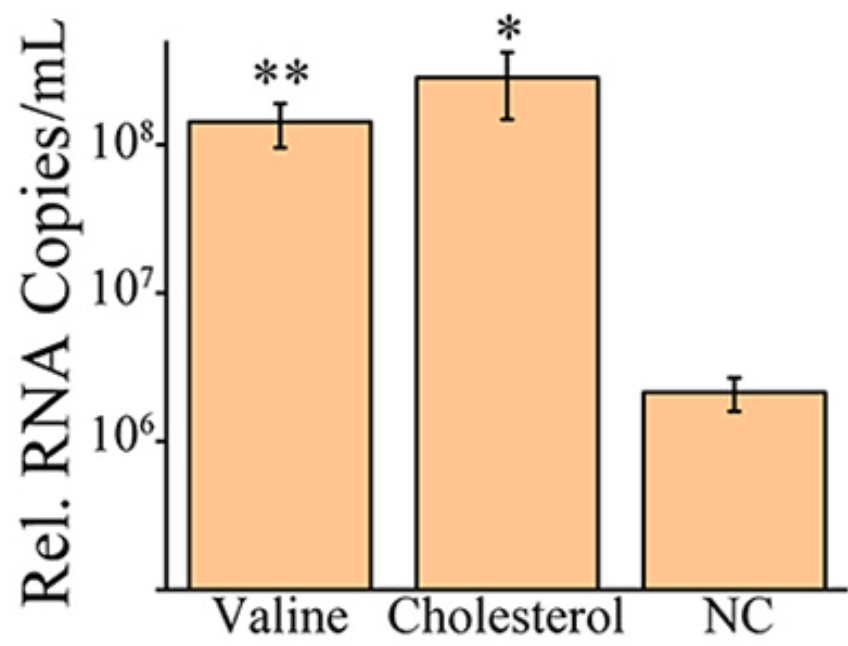
Coronaviruses are commonly characterized by a unique discontinuous RNA transcriptional synthesis strategy guided by transcription-regulating sequences (TRSs). However, the details of RNA synthesis in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have not been fully elucidated. Here, we present a timescaled, gene-comparable transcriptome of SARS-CoV-2, demonstrating that ACGAAC functions as a core TRS guiding the discontinuous RNA synthesis of SARS-CoV-2 from a holistic perspective. During infection, viral transcription, rather than genome replication, dominates all viral RNA synthesis activities. The most highly expressed viral gene is the nucleocapsid gene, followed by ORF7 and ORF3 genes, while the envelope gene shows the lowest expression. Host transcription dysregulation keeps exacerbating after viral RNA synthesis reaches a maximum. The most enriched host pathways are metabolism related. Two of them (cholesterol and valine metabolism) affect viral replication in reverse. Furthermore, the activation of numerous cytokines emerges before large-scale viral RNA synthesis. IMPORTANCE SARS-CoV-2 is responsible for the current severe global health emergency that began at the end of 2019. Although the universal transcriptional strategies of coronaviruses are preliminarily understood, the details of RNA synthesis, especially the timematched transcription level of each SARS-CoV-2 gene and the principles of subgenomic mRNA synthesis, are not clear. The coterminal subgenomic mRNAs of SARS-CoV-2 present obstacles in identifying the expression of most genes by PCR-based methods, which are exacerbated by the lack of related antibodies. Moreover, SARS-CoV-2-related metabolic imbalance and cytokine storm are receiving increasing attention from both clinical and mechanistic perspectives. Our transcriptomic research provides information on both viral RNA synthesis and host responses, in which the transcription-regulating sequences and transcription levels of viral genes are demonstrated, and the metabolic dysregulation and cytokine levels identified at the host cellular level support the development of novel medical treatment strategies.
were selected and referred to as short "query reads" (see Fig. S1 ). Their first nt were located one by one downstream at a specific region of the 59 UTR. By querying the combined read pool containing all viral sequences, all sequences of 30 nt in length whose 59 15-nt sequences were identical to the query reads were returned and their numbers were counted. The 15-nt sequences downstream of the corresponding query reads were referred to as their "return reads." Located around the possible TRS, the return reads could be either manually aligned continuously to gRNAs or aligned discontinuously to sgmRNAs with the 59 partial sequence homologous to the leader UTR (upstream of the leader TRS) and the 39 sequence homologous to various ORFs (downstream of the body TRS). When a site could be regarded as either 59 continuous (continuous to upstream query reads) or 39 continuous (continuous to downstream ORFs), it was designated 59 continuous, as we intended to identify the probable leader TRS as long as possible. In parallel, two additional types of 15-nt query reads with sequences homologous to the beginning of the ORFs were used: the first type started at the body TRS (6 nt) and ended at 19 nt of the downstream ORF (referred to as "in-TRS reads"), and the second type was homologous to 11 to 115 nt of the ORFs adjacent to the downstream body TRS (referred to as "after-TRS reads"). For each known ORF, an in-TRS read and an after-TRS read were..
References
Blanco-Melo, Nilsson-Payant, Liu, Uhl, Hoagland et al., Imbalanced host response to SARS-CoV-2 drives development of COVID-19, Cell,
doi:10.1016/j.cell.2020.04.026Bojkova, Klann, Koch, Widera, Krause et al., Proteomics of SARS-CoV-2-infected host cells reveals therapy targets, Nature,
doi:10.1038/s41586-020-2332-7Chua, Lukassen, Trump, Hennig, Wendisch et al., COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis, Nat Biotechnol,
doi:10.1038/s41587-020-0602-4Daniloski, Jordan, Wessels, Hoagland, Kasela et al., Identification of required host factors for SARS-CoV-2 infection in human cells, Cell,
doi:10.1016/j.cell.2020.10.030Hachim, Kavian, Cohen, Chin, Chu et al., ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection, Nat Immunol,
doi:10.1038/s41590-020-0773-7Hoffmann, Kleine-Weber, Schroeder, Krüger, Herrler et al., SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor, Cell,
doi:10.1016/j.cell.2020.02.052Lee, Cao, Mostoslavsky, Lombard, Liu et al., A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy, Proc Natl Acad Sci U S A,
doi:10.1073/pnas.0712145105Lee, Park, Jeong, Ahn, Choi et al., Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19, Sci Immunol,
doi:10.1126/sciimmunol.abd1554Liao, Liu, Yuan, Wen, Xu et al., Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19, Nat Med,
doi:10.1038/s41591-020-0901-9Livak, Schmittgen, Analysis of relative gene expression data using real-time quantitative PCR and the 22DDCT method, Methods,
doi:10.1006/meth.2001.1262Miyazaki, Ichiki, Hashimoto, Inanaga, Imayama et al., SIRT1, a longevity gene, downregulates angiotensin II type 1 receptor expression in vascular smooth muscle cells, Arterioscler Thromb Vasc Biol,
doi:10.1161/ATVBAHA.108.166991Radenkovic, Chawla, Pirro, Sahebkar, Banach, Cholesterol in relation to COVID-19: should we care about it?, J Clin Med,
doi:10.3390/jcm9061909Salomon, Hoffmann, Webster, Inhibition of the cytokine response does not protect against lethal H5N1 influenza infection, Proc Natl Acad Sci U S A,
doi:10.1073/pnas.0705289104Snijder, Bredenbeek, Dobbe, Thiel, Ziebuhr et al., Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage, J Mol Biol,
doi:10.1016/S0022-2836(03)00865-9Snijder, Decroly, Ziebuhr, Chapter three -the nonstructural proteins directing coronavirus RNA synthesis and processing
Storz, Forkhead homeobox type O transcription factors in the responses to oxidative stress, Antioxid Redox Signal,
doi:10.1089/ars.2010.3405Thiel, Ivanov, Putics, Hertzig, Schelle et al., Mechanisms and enzymes involved in SARS coronavirus genome expression, J Gen Virol,
doi:10.1099/vir.0.19424-0Wang, Hu, Hu, Zhu, Liu et al., Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China, JAMA,
doi:10.1001/jama.2020.1585Wang, Simoneau, Kulsuptrakul, Bouhaddou, Travisano et al., Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses, Cell,
doi:10.1016/j.cell.2020.12.004Xiong, Liu, Cao, Wang, Guo et al., Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients, Emerg Microbes Infect,
doi:10.1080/22221751.2020.1747363Yao, Irwin, Zhao, Nilsen, Hamilton et al., Mitochondrial bioenergetic deficit precedes Alzheimer's pathology in female mouse model of Alzheimer's disease, Proc Natl Acad Sci U S A,
doi:10.1073/pnas.0903563106Zhang, Li, Cruz, Kone, Sirtuin 1 functionally and physically interacts with disruptor of telomeric silencing-1 to regulate alpha-ENaC transcription in collecting duct, J Biol Chem,
doi:10.1074/jbc.M109.020073DOI record:
{
"DOI": "10.1128/jvi.00600-21",
"ISSN": [
"0022-538X",
"1098-5514"
],
"URL": "http://dx.doi.org/10.1128/jvi.00600-21",
"abstract": "<jats:p>SARS-CoV-2 is responsible for the current severe global health emergency that began at the end of 2019. Although the universal transcriptional strategies of coronaviruses are preliminarily understood, the details of RNA synthesis, especially the time-matched transcription level of each SARS-CoV-2 gene and the principles of subgenomic mRNA synthesis, are not clear.</jats:p>",
"alternative-id": [
"10.1128/JVI.00600-21"
],
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2021-04-07"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 1,
"value": "2021-06-02"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 2,
"value": "2021-08-25"
}
],
"author": [
{
"ORCID": "http://orcid.org/0000-0002-3974-3421",
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"authenticated-orcid": true,
"family": "Wang",
"given": "Xuefeng",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
},
{
"name": "School of Life Sciences, Northeast Normal University, Changchun, People’s Republic of China"
}
],
"family": "Zhao",
"given": "Yudong",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"family": "Yan",
"given": "Feihu",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"family": "Wang",
"given": "Tiecheng",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"family": "Sun",
"given": "Weiyang",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"family": "Feng",
"given": "Na",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
},
{
"name": "Key Laboratory of Animal Resistant Biology of Shandong, College of Life Sciences, Shandong Normal University, Jinan, People’s Republic of China"
}
],
"family": "Wang",
"given": "Wenqi",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Key Laboratory of Animal Resistant Biology of Shandong, College of Life Sciences, Shandong Normal University, Jinan, People’s Republic of China"
}
],
"family": "Wang",
"given": "Hongmei",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-7438-0638",
"affiliation": [
{
"name": "Key Laboratory of Animal Resistant Biology of Shandong, College of Life Sciences, Shandong Normal University, Jinan, People’s Republic of China"
}
],
"authenticated-orcid": true,
"family": "He",
"given": "Hongbin",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"family": "Yang",
"given": "Songtao",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"family": "Xia",
"given": "Xianzhu",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, People’s Republic of China"
}
],
"family": "Gao",
"given": "Yuwei",
"sequence": "additional"
}
],
"container-title": "Journal of Virology",
"container-title-short": "J Virol",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"journals.asm.org"
]
},
"created": {
"date-parts": [
[
2021,
6,
13
]
],
"date-time": "2021-06-13T23:05:56Z",
"timestamp": 1623625556000
},
"deposited": {
"date-parts": [
[
2022,
3,
5
]
],
"date-time": "2022-03-05T16:28:34Z",
"timestamp": 1646497714000
},
"editor": [
{
"affiliation": [],
"family": "Subbarao",
"given": "Kanta",
"sequence": "additional"
}
],
"funder": [
{
"DOI": "10.13039/501100012166",
"award": [
"2016YFD0500203"
],
"doi-asserted-by": "publisher",
"name": "National Key Research and Development Program of China"
}
],
"indexed": {
"date-parts": [
[
2024,
2,
21
]
],
"date-time": "2024-02-21T11:09:16Z",
"timestamp": 1708513756036
},
"is-referenced-by-count": 8,
"issue": "18",
"issued": {
"date-parts": [
[
2021,
8,
25
]
]
},
"journal-issue": {
"issue": "18",
"published-print": {
"date-parts": [
[
2021,
8,
25
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://doi.org/10.1128/ASMCopyrightv2",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
8,
25
]
],
"date-time": "2021-08-25T00:00:00Z",
"timestamp": 1629849600000
}
},
{
"URL": "https://journals.asm.org/non-commercial-tdm-license",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
8,
25
]
],
"date-time": "2021-08-25T00:00:00Z",
"timestamp": 1629849600000
}
}
],
"link": [
{
"URL": "https://journals.asm.org/doi/pdf/10.1128/JVI.00600-21",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://journals.asm.org/doi/pdf/10.1128/JVI.00600-21",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "235",
"original-title": [],
"prefix": "10.1128",
"published": {
"date-parts": [
[
2021,
8,
25
]
]
},
"published-print": {
"date-parts": [
[
2021,
8,
25
]
]
},
"publisher": "American Society for Microbiology",
"reference": [
{
"key": "e_1_3_3_2_2",
"unstructured": "World Health Organization. 2020. WHO coronavirus disease (COVID-19) dashboard. https://covid19.who.int/."
},
{
"DOI": "10.1038/s41586-020-2008-3",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_3_2"
},
{
"author": "Snijder EJ",
"first-page": "59",
"key": "e_1_3_3_4_2",
"unstructured": "Snijder EJ, Decroly E, Ziebuhr J. 2016. Chapter three - the nonstructural proteins directing coronavirus RNA synthesis and processing, p 59–126. In Ziebuhr J (ed), Advances in virus research, vol 96. Academic Press.",
"volume-title": "Advances in virus research",
"year": "2016"
},
{
"DOI": "10.1016/S0022-2836(03)00865-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_5_2"
},
{
"DOI": "10.1146/annurev-virology-100114-055218",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_6_2"
},
{
"DOI": "10.1016/j.cell.2020.04.011",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_7_2"
},
{
"DOI": "10.1099/vir.0.19424-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_8_2"
},
{
"DOI": "10.1101/2020.03.05.976167",
"doi-asserted-by": "crossref",
"key": "e_1_3_3_9_2",
"unstructured": "Taiaroa G Rawlinson D Featherstone L Pitt M Caly L Druce J Purcell D Harty L Tran T Roberts J Scott N Catton M Williamson D Coin L Duchene S. 2020. Direct RNA sequencing and early evolution of SARS-CoV-2. bioRxiv 10.1101/2020.03.05.976167:2020.03.05.976167."
},
{
"DOI": "10.1038/s41587-020-0602-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_10_2"
},
{
"DOI": "10.1080/22221751.2020.1747363",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_11_2"
},
{
"DOI": "10.1101/2020.06.17.156455",
"doi-asserted-by": "crossref",
"key": "e_1_3_3_12_2",
"unstructured": "Stukalov A Girault V Grass V Bergant V Karayel O Urban C Haas DA Huang Y Oubraham L Wang A Hamad SM Piras A Tanzer M Hansen FM Enghleitner T Reinecke M Lavacca TM Ehmann R Wölfel R Jores J Kuster B Protzer U RR Ziebuhr J Thiel V Scaturro P Mann M Pichlmair A. 2020. Multi-level proteomics reveals host-perturbation strategies of SARS-CoV-2 and SARS-CoV. bioRxiv 10.1101/2020.06.17.156455:2020.06.17.156455."
},
{
"DOI": "10.1073/pnas.0903563106",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_13_2"
},
{
"DOI": "10.1038/s41586-020-2332-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_14_2"
},
{
"DOI": "10.1016/j.tem.2013.12.001",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_15_2"
},
{
"DOI": "10.1016/j.molcel.2007.07.032",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_16_2"
},
{
"DOI": "10.1074/jbc.M109.020073",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_17_2"
},
{
"DOI": "10.1089/ars.2010.3405",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_18_2"
},
{
"DOI": "10.1073/pnas.0712145105",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_19_2"
},
{
"DOI": "10.1016/j.cell.2020.02.052",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_20_2"
},
{
"DOI": "10.1001/jama.2020.1585",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_21_2"
},
{
"DOI": "10.1161/ATVBAHA.108.166991",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_22_2"
},
{
"DOI": "10.1146/annurev-physiol-020518-114455",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_23_2"
},
{
"DOI": "10.1038/sj.onc.1202568",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_24_2"
},
{
"DOI": "10.1038/s41591-020-0901-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_25_2"
},
{
"DOI": "10.1038/s41590-020-0773-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_26_2"
},
{
"DOI": "10.1016/j.cell.2020.12.004",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_27_2"
},
{
"DOI": "10.1016/j.cell.2020.10.030",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_28_2"
},
{
"DOI": "10.3390/jcm9061909",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_29_2"
},
{
"DOI": "10.1080/14728222.2019.1586886",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_30_2"
},
{
"DOI": "10.1126/sciimmunol.abd1554",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_31_2"
},
{
"DOI": "10.1016/j.cell.2020.04.026",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_32_2"
},
{
"DOI": "10.1073/pnas.0705289104",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_33_2"
},
{
"DOI": "10.1006/meth.2001.1262",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_34_2"
}
],
"reference-count": 33,
"references-count": 33,
"relation": {},
"resource": {
"primary": {
"URL": "https://journals.asm.org/doi/10.1128/JVI.00600-21"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Virology",
"Insect Science",
"Immunology",
"Microbiology"
],
"subtitle": [],
"title": "Viral and Host Transcriptomes in SARS-CoV-2-Infected Human Lung Cells",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1128/asmj-crossmark-policy-page",
"volume": "95"
}