Are Non-Structural Proteins From SARS-CoV-2 the Target of Hydroxychloroquine? An in Silico Study
E F Guimarães, Ériky Fernandes, Guimarães Silva, Bruna Fernandes, Gabriel Luan, Angélica Pinto, Fátima De, Anderson Dillmann Marcussi, Groto, Nayara Kádima, Teixeira
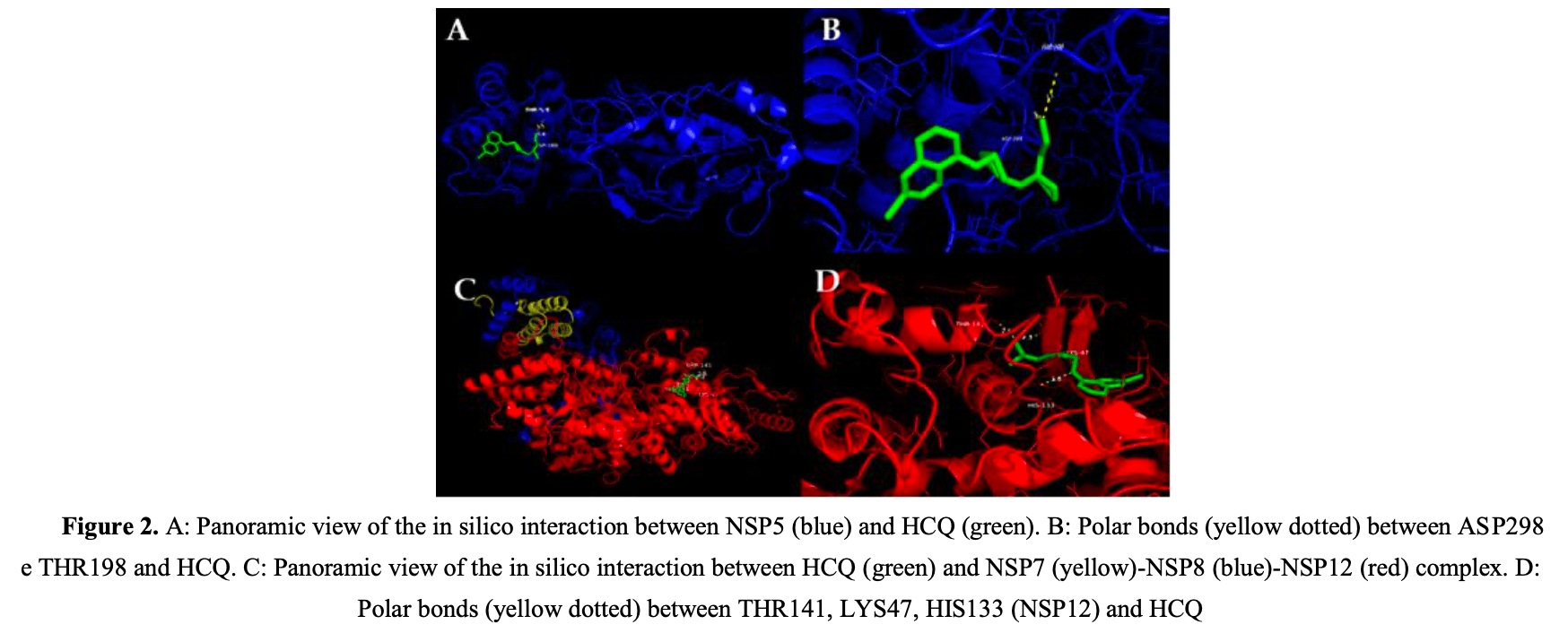
COVID-19 is caused by SARS-CoV-2 which has structural and non-structural proteins (NSP) essential for infection and viral replication. There is a possible binding of SARS-CoV-2 to the beta-1 chain of hemoglobin in red blood cells and thus, decreasing the oxygen transport capacity. Since hydroxychloroquine (HCQ) can accumulate in red cells, there is a chance of interaction of this drug with the virus. To analyze possible interactions between SARS-CoV-2 NSP and hemoglobin with the HCQ using molecular docking and implications for the infected host. This research consisted of a study using bioinformatics tools. The files of the protein structures and HCQ were prepared using the AutoDock Tools software. These files were used to perform molecular docking simulations by AutoDock Vina. The binding affinity report of the generated conformers was analyzed using PyMol software, as well as the chemical bonds formed. The results showed that HCQ is capable of interacting with both SARS-CoV-2 NSP and human hemoglobin. The HCQ/NSP3 conformer, HCQ/NSP5, HCQ/NSP7-NSP8-NSP12, HCQ/NSP9, HCQ/NSP10-NSP16 showed binding affinity. In addition, the interaction between HCQ and hemoglobin resulted in polar bonds. Interaction between SARS-CoV-2 NSP and HCQ indicates that this drug possibly acts by preventing the continuity of infection.
Acta Medica Iranica, Vol. 61, No. 2 (2023) 103 on NSP, the therapeutic action of HCQ could not be effective since mechanisms such as cytokine storm could have already been activated by the course of infection.
References
Angeletti, Benvenuto, Bianchi, Giovanetti, Pascarella et al., COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis, J Med Virol
Ben-Zvi, Kivity, Langevitz, Shoenfeld, Hydroxychloroquine: from malaria to autoimmunity, Clin Rev Allergy Immunol
Cavezzi, Troiani, Corrao, COVID-19: hemoglobin, iron, and hypoxia beyond inflammation. A narrative review, Clin Pract
Chen, Fang, Tam, Liu, Formation of stable homodimer via the C-terminal α-helical domain of coronavirus nonstructural protein 9 is critical for its function in viral replication, Virology
Chen, Su, Ke, Xu, Zhang, Biochemical and Structural Insights into the Mechanisms of SARS Coronavirus RNA Ribose 2′-O-Methylation by nsp16/nsp10 Protein Complex, PLoS Pathog
Claverie, A Putative Role of de-Mono-ADP-Ribosylation of STAT1 by the SARS-CoV-2 Nsp3 Protein in the Cytokine Storm Syndrome of COVID-19, Viruses
Da Silva, Figueiredo, Byler, Setzer, Essential Oils as Antiviral Agents, Potential of Essential Oils to Treat SARS-CoV-2 Infection: An In-Silico Investigation, Int J Mol Sci
Deshpande, Tiwari, Nyayanit, Modak, In silico molecular docking analysis for repurposing therapeutics against multiple proteins from SARS-CoV-2, Eur J Pharmacol
Egloff, Ferron, Campanacci, Longhi, Rancurel et al., The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a singlestranded RNA-binding subunit unique in the RNA virus world, Proc Natl Acad Sci
Encinar, Menendez, Potential Drugs Targeting Early Innate Immune Evasion of SARS-Coronavirus 2 via 2'-O-Methylation of Viral RNA, Viruses
Fehr, Channappanavar, Jankevicius, Fett, Zhao et al., The Conserved Coronavirus Macrodomain Promotes Virulence and Suppresses the Innate Immune Response during Severe Acute Respiratory Syndrome Coronavirus Infection, mBio
Ferrari, Cutler, Kinetics and thermodynamics of chloroquine and hydroxychloroquine transport across the human erythrocyte membrane, Biochem Pharmacol
Gordon, Jang, Bouhaddou, Xu, Obernier et al., A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature
He, Deng, Li, Coronavirus disease 2019: What we know?, J Med Virol
Kaushal, Gupta, Goyal, Khaiboullina, Baranwal et al., Mutational Frequencies of SARS-CoV-2 Genome during the Beginning Months of the Outbreak in USA, Pathogens
Khailany, Safdar, Ozaslan, Genomic characterization of a novel SARS-CoV-2, Gene Rep
Kirchdoerfer, Ward, Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors, Nat Commun
Leach, The Use of Molecular Modelling and Chemoinformatics to Discover and Design New Molecules
Liu, Wei, Li, Yang, Tan, Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2, J Med Virol
Malik, Properties of Coronavirus and SARS-CoV-2, Malays J Pathol
Maurya, Maurya, Mishra, Siddique, Virtual screening, ADME/T, and binding free energy analysis of anti-viral, anti-protease, and anti-infectious compounds against NSP10/NSP16 methyltransferase and main protease of SARS CoV-2, J Recept Signal Transduct Res
Miknis, Donaldson, Umland, Rimmer, Baric et al., Severe Acute Respiratory Syndrome Coronavirus nsp9 Dimerization Is Essential for Efficient Viral Growth, J Virol
Morris, Lim-Wilby, Molecular Docking, Methods Mol Biol
Pastick, Okafor, Wang, Lofgren, Skipper et al., Review: Hydroxychloroquine and Chloroquine for Treatment of SARS-CoV-2 (COVID-19), Open Forum Infect Dis
Quimque, Notarte, Fernandez, Mendoza, Liman et al., Virtual screeningdriven drug discovery of SARS-CoV2 enzyme inhibitors targeting viral attachment, replication, post-translational modification and host immunity evasion infection mechanisms, J Biomol Struct Dyn
Rosas-Lemus, Minasov, Shuvalova, Inniss, Kiryukhina et al., The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine, bioRxiv
Shah, Modi, Sagar, In silico studies on therapeutic agents for COVID-19: Drug repurposing approach, Life Sci
Shityakov, Förster, In silico predictive model to determine vector-mediated transport properties for the blood-brain barrier choline transporter, Adv Appl Bioinform Chem
Sinha, Balayla, Hydroxychloroquine and covid-19, Postgraduate Med J
Snijder, Decroly, Ziebuhr, The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing, Adv Virus Res
Subissi, Imbert, Ferron, Collet, Coutard et al., SARS-CoV ORF1b-encoded nonstructural proteins 12-16: Replicative enzymes as antiviral targets, Antiviral Res
Subissi, Posthuma, Collet, Zevenhoven-Dobbe, Gorbalenya et al., One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities, Proc Natl Acad Sci
Tahir Ul Qamar, Alqahtani, Alamri, Chen, Structural basis of SARS-CoV-2 3CLpro and anti-COVID-SARS-CoV-2 proteins as hydroxychloroquine targets 104, Acta Medica Iranica
Thomas-Rüddel, Winning, Dickmann, Ouart, Kortgen et al., Coronavirus disease 2019 (COVID-19): update for anesthesiologists and intensivists March 2020, Anaesthesist
Trott, Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J Comput Chem
Velavan, Meyer, The COVID-19 epidemic, Trop Med Int Heal
Velthuis, Van Den Worm, She, Sims, Baric et al., Zn2+ Inhibits Coronavirus and Arterivirus RNA Polymerase Activity In Vitro and Zinc Ionophores Block the Replication of These Viruses in Cell Culture, PLoS Pathog
Wang, Horby, Hayden, Gao, A novel coronavirus outbreak of global health concern, Lancet
Wellems, Malaria, How chloroquine works, Nature
Wenzhong, Hualan, COVID-19: Attacks the 1-Beta Chain of Hemoglobin and Captures the Porphyrin to Inhibit Human Heme Metabolism, ChemRxiv
Wu, Peng, Huang, Ding, Niu et al., Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China, Cell Host Microbe
Wu, Zhao, Yu, Chen, Song, A new coronavirus associated with human respiratory disease in China, Nature
Xue, Moyer, Peng, Wu, Hannafon et al., Chloroquine Is a Zinc Ionophore, PLoS One
Yoshimoto, The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19, Protein J
Yuen, Lam, Wong, Mak, Wang et al., SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists, Emerg Microbes Infect
Zhai, Sun, Li, Pang, Xu et al., Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer, Nat Struct Mol Biol
Zhang, Lin, Sun, Curth, Drosten et al., Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors, Science
Zhao, Liu, Yu, Wang, Du et al., The characteristics and clinical value of chest CT images of novel coronavirus pneumonia, Clin Radiol
Zhou, Yu, Du, Fan, Liu et al., Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study, Lancet
Zhu, Chen, Tian, Zhou, Xu et al., Porcine Deltacoronavirus 1 nsp5 Cleaves DCP1A to Decrease Its Antiviral Ability, J Virol
Zhu, Zhang, Li, Yang, Song, A Novel Coronavirus from Patients with Pneumonia in China, 2019, N Engl J Med
DOI record:
{
"DOI": "10.18502/acta.v61i2.12533",
"ISSN": [
"1735-9694",
"0044-6025"
],
"URL": "http://dx.doi.org/10.18502/acta.v61i2.12533",
"abstract": "<jats:p>COVID-19 is caused by SARS-CoV-2 which has structural and non-structural proteins (NSP) essential for infection and viral replication. There is a possible binding of SARS-CoV-2 to the beta-1 chain of hemoglobin in red blood cells and thus, decreasing the oxygen transport capacity. Since hydroxychloroquine (HCQ) can accumulate in red cells, there is a chance of interaction of this drug with the virus. To analyze possible interactions between SARS-CoV-2 NSP and hemoglobin with the HCQ using molecular docking and implications for the infected host. This research consisted of a study using bioinformatics tools. The files of the protein structures and HCQ were prepared using the AutoDock Tools software. These files were used to perform molecular docking simulations by AutoDock Vina. The binding affinity report of the generated conformers was analyzed using PyMol software, as well as the chemical bonds formed. The results showed that HCQ is capable of interacting with both SARS-CoV-2 NSP and human hemoglobin. The HCQ/NSP3 conformer, HCQ/NSP5, HCQ/NSP7-NSP8-NSP12, HCQ/NSP9, HCQ/NSP10-NSP16 showed binding affinity. In addition, the interaction between HCQ and hemoglobin resulted in polar bonds. Interaction between SARS-CoV-2 NSP and HCQ indicates that this drug possibly acts by preventing the continuity of infection.</jats:p>",
"author": [
{
"affiliation": [],
"family": "Guimarães Silva",
"given": "Ériky Fernandes",
"sequence": "first"
},
{
"affiliation": [],
"family": "Fernandes",
"given": "Bruna",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Pinto",
"given": "Luan Gabriel",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Marcuss",
"given": "Angélica De Fátima",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Dillmann Groto",
"given": "Anderson",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Nayara Teixeira",
"given": "Kádima",
"sequence": "additional"
}
],
"container-title": "ACTA MEDICA IRANICA",
"container-title-short": "ACTA",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2023,
4,
24
]
],
"date-time": "2023-04-24T08:24:27Z",
"timestamp": 1682324667000
},
"deposited": {
"date-parts": [
[
2023,
10,
21
]
],
"date-time": "2023-10-21T16:58:20Z",
"timestamp": 1697907500000
},
"indexed": {
"date-parts": [
[
2024,
9,
16
]
],
"date-time": "2024-09-16T19:38:46Z",
"timestamp": 1726515526593
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2023,
4,
18
]
]
},
"link": [
{
"URL": "https://publish.kne-publishing.com/index.php/ACTA/article/download/12533/11887",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://publish.kne-publishing.com/index.php/ACTA/article/download/12533/11887",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "7770",
"original-title": [],
"prefix": "10.18502",
"published": {
"date-parts": [
[
2023,
4,
18
]
]
},
"published-online": {
"date-parts": [
[
2023,
4,
18
]
]
},
"publisher": "Knowledge E DMCC",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://publish.kne-publishing.com/index.php/ACTA/article/view/12533"
},
"secondary": [
{
"URL": "https://acta.tums.ac.ir/index.php/acta/article/view/9090",
"label": "UNIVERSITY"
},
{
"URL": "https://publish.kne-publishing.com/index.php/ACTA/article/view/12533",
"label": "PUBLISHER"
}
]
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Are Non-Structural Proteins From SARS-CoV-2 the Target of Hydroxychloroquine? An in Silico Study",
"type": "journal-article"
}