Human SARS-CoV-2 challenge uncovers local and systemic response dynamics
Rik G H Lindeboom, Kaylee B Worlock, Lisa M Dratva, Masahiro Yoshida, David Scobie, Helen R Wagstaffe, Laura Richardson, Anna Wilbrey-Clark, Josephine L Barnes, Lorenz Kretschmer, Krzysztof Polanski, Jessica Allen-Hyttinen, Puja Mehta, Dinithi Sumanaweera, Jacqueline M Boccacino, Waradon Sungnak, Rasa Elmentaite, Ni Huang, Lira Mamanova, Rakesh Kapuge, Liam Bolt, Elena Prigmore, Ben Killingley, Mariya Kalinova, Maria Mayer, Alison Boyers, Alex Mann, Leo Swadling, Maximillian N J Woodall, Samuel Ellis, Claire M Smith, Vitor H Teixeira, Sam M Janes, Rachel C Chambers, Muzlifah Haniffa, Andrew Catchpole, Robert Heyderman, Mahdad Noursadeghi, Benny Chain, Andreas Mayer, Kerstin B Meyer, Christopher Chiu, Marko Z Nikolić, Sarah A Teichmann
Nature, doi:10.1038/s41586-024-07575-x
bias as participation was voluntary and instigated by the volunteers. Due to these factors, direct extrapolation of the results to young children, older adults, those with pre-existing conditions and minority groups may not be possible.
Article Sanger Institute through the Sanger Prize. This publication is part of the Human Cell Atlas (https://www.humancellatlas.org/publications). Illustrations in Fig. 1 and Extended Data Fig. 10 were created using BioRender (https://www.biorender.com). For the purpose of Open Access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission. Author contributions M.Z.N. and S.A.T. conceived, set up, directed this study and provided funding. C.C. set up the clinical study and co-ordinated sampling. K.B.W. optimized digestion protocols, processed samples for 10x and CITE-seq, isolated DNA for genotyping, performed Dextramer experiments and assisted with data analyses and interpretation. R.G.H.L. performed and led the data analyses. L.M.D. assisted with data analyses and implemented Cell2TCR in Python. L.K., J.M.B., R.E., K.P., W.S., N.H. and D. Sumanaweera advised on and assisted with data analyses. R.G.H.L., K.B.W., L.M.D., K.B.M., M.Z.N. and S.A.T. interpreted the data and wrote the manuscript. L.R., A.W.-C., L.M., R.K., L.B. and E.P. performed the single-cell sequencing library preparations. M.Y., J.L.B. and J.A.-H. assisted with CITE-seq and 10x sample processing. V.H.T., S.M.J. and R.C.C. provided student supervision to K.B.W. and P.M. H.R.W. processed blood samples. D. Scobie, B.C. and A. Mayer provided bulk TCR-seq data and advised on the data analysis. P.M. collected nasopharyngeal samples for..
References
Altman, Phenotypic analysis of antigen-specific T lymphocytes, Science
Augusto, Healthcare Workers Bioresource: study outline and baseline characteristics of a prospective healthcare worker cohort to study immune protection and pathogenesis in COVID-19, Wellcome Open Res
Bingham, Pyro: deep universal probabilistic programming, J. Mach. Learn. Res
Blanco-Melo, Imbalanced host response to SARS-CoV-2 drives development of COVID-19, Cell
Bonilla, Chai, Williams, Multi-task Gaussian process prediction
Chandran, Rapid synchronous type 1 IFN and virus-specific T cell responses characterize first wave non-severe SARS-CoV-2 infections, Cell Rep. Med
Dan, A cytokine-independent approach to identify antigen-specific human germinal center T follicular helper cells and rare antigen-specific CD4 + T cells in blood, J. Immunol
Domínguez Conde, Cross-tissue immune cell analysis reveals tissue-specific features in humans, Science
Fears, The dynamics of γδ T cell responses in nonhuman primates during SARS-CoV-2 infection, Commun. Biol
Frere, SARS-CoV-2 infection in hamsters and humans results in lasting and unique systemic perturbations after recovery, Sci. Transl. Med
Gardner, Pleiss, Weinberger, Bindel, Wilson et al., blackbox matrix-matrix gaussian process inference with GPU acceleration
Hadjadj, Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients, Science
Heaton, Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes, Nat. Methods
Kaneko, Temporal changes in T cell subsets and expansion of cytotoxic CD4 T cells in the lungs in severe COVID-19, Clin. Immunol,
doi:10.1016/j.clim.2022.108991Killingley, Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults, Nat. Med
Korsunsky, Fast, sensitive and accurate integration of single-cell data with Harmony, Nat. Methods
Lenormand, HLA-DQA2 and HLA-DQB2 genes are specifically expressed in human Langerhans cells and encode a new HLA class II molecule, J. Immunol
Liu, Time-resolved systems immunology reveals a late juncture linked to fatal COVID-19, Cell
Loske, Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children, Nat. Biotechnol
Love, Huber, Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol
Luecken, Theis, Current best practices in single-cell RNA-seq analysis: a tutorial, Mol. Syst. Biol
Madissoon, A spatially resolved atlas of the human lung characterizes a gland-associated immune niche, Nat. Genet
Mayer, Callan, Measures of epitope binding degeneracy from T cell receptor repertoires, Proc. Natl Acad. Sci. USA
Mayer-Blackwell, TCR meta-clonotypes for biomarker discovery with enabled identification of public, HLA-restricted clusters of SARS-CoV-2 TCRs, eLife
Meckiff, Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4 T cells in COVID-19, Cell
Oakes, Quantitative characterization of the T cell receptor repertoire of naïve and memory subsets using an integrated experimental and computational pipeline which is robust, economical, and versatile, Front. Immunol
Peacock, Heather, Ronel, Chain, Decombinator V4: an improved AIRR compliant-software package for T-cell receptor sequence annotation?, Bioinformatics
Policard, Jain, Rego, Dakshanamurthy, Immune characterization and profiles of SARS-CoV-2 infected patients reveals potential host therapeutic targets and SARS-CoV-2 oncogenesis mechanism, Virus Res
Ren, COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas, Cell
Rosenheim, SARS-CoV-2 human challenge reveals single-gene blood transcriptional biomarkers that discriminate early and late phases of acute respiratory viral infections,
doi:10.1101/2023.06.01.23290819Rudy, Lew, The nonpolymorphic MHC class II isotype, HLA-DQA2, is expressed on the surface of B lymphoblastoid cells, J. Immunol
Saichi, Single-cell RNA sequencing of blood antigen-presenting cells in severe COVID-19 reveals multi-process defects in antiviral immunity, Nat. Cell Biol
Schulte-Schrepping, Severe COVID-19 is marked by a dysregulated myeloid cell compartment, Cell
Shen, ACE2-independent infection of T lymphocytes by SARS-CoV-2, Signal Transduct. Target. Ther
Shenoy, Antigen presentation by lung epithelial cells directs CD4 + TRM cell function and regulates barrier immunity, Nat. Commun
Singanayagam, Community transmission and viral load kinetics of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study, Lancet Infect. Dis
Stephenson, Single-cell multi-omics analysis of the immune response in COVID-19, Nat. Med
Sturm, Scirpy: a Scanpy extension for analyzing single-cell T-cell receptor-sequencing data, Bioinformatics
Suo, Dandelion uses the single-cell adaptive immune receptor repertoire to explore lymphocyte developmental origins, Nat. Biotechnol
Suo, Mapping the developing human immune system across organs, Science
Tareen, Kinney, Logomaker: beautiful sequence logos in Python, Bioinformatics
Titsias, Lawrence, Bayesian Gaussian process latent variable model, Proc. Mach. Learn. Res
Traag, Waltman, Van Eck, From Louvain to Leiden: guaranteeing well-connected communities, Sci. Rep
Uddin, An economical, quantitative, and robust protocol for high-throughput T cell receptor sequencing from tumor or blood, Methods Mol. Biol,
doi:10.1007/978-1-4939-8885-3_2Wagih, ggseqlogo: a versatile R package for drawing sequence logos, Bioinformatics
Wagstaffe, Mucosal and systemic immune correlates of viral control after SARS-CoV-2 infection challenge in seronegative adults, Sci. Immunol
Wang, CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells, Signal Transduct. Target. Ther
Wigerblad, Single-cell analysis reveals the range of transcriptional states of circulating human neutrophils, J. Immunol
Wilkinson, Preexisting influenza-specific CD4 + T cells correlate with disease protection against influenza challenge in humans, Nat. Med
Wolf, Angerer, Theis, SCANPY: large-scale single-cell gene expression data analysis, Genome Biol
Woodall, Age-specific nasal epithelial responses to SARS-CoV-2 infection, Nat. Microbiol
Woodall, Masonou, Case, Smith, Human models for COVID-19 research, J. Physiol
Wosen, Mukhopadhyay, Macaubas, Mellins, Epithelial MHC class II expression and its role in antigen presentation in the gastrointestinal and respiratory tracts, Front. Immunol
Yoshida, Local and systemic responses to SARS-CoV-2 infection in children and adults, Nature
Young, Behjati, SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data, Gigascience
Ziegler, Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19, Cell
{ 'indexed': {'date-parts': [[2024, 6, 21]], 'date-time': '2024-06-21T17:36:21Z', 'timestamp': 1718991381661},
'reference-count': 63,
'publisher': 'Springer Science and Business Media LLC',
'license': [ { 'start': { 'date-parts': [[2024, 6, 19]],
'date-time': '2024-06-19T00:00:00Z',
'timestamp': 1718755200000},
'content-version': 'tdm',
'delay-in-days': 0,
'URL': 'https://creativecommons.org/licenses/by/4.0'},
{ 'start': { 'date-parts': [[2024, 6, 19]],
'date-time': '2024-06-19T00:00:00Z',
'timestamp': 1718755200000},
'content-version': 'vor',
'delay-in-days': 0,
'URL': 'https://creativecommons.org/licenses/by/4.0'}],
'content-domain': {'domain': ['link.springer.com'], 'crossmark-restriction': False},
'abstract': '<jats:title>Abstract</jats:title><jats:p>The COVID-19 pandemic is an ongoing global health '
'threat, yet our understanding of the dynamics of early cellular responses to this disease '
'remains limited<jats:sup>1</jats:sup>. Here in our SARS-CoV-2 human challenge study, we used '
'single-cell multi-omics profiling of nasopharyngeal swabs and blood to temporally resolve '
'abortive, transient and sustained infections in seronegative individuals challenged with '
'pre-Alpha SARS-CoV-2. Our analyses revealed rapid changes in cell-type proportions and dozens '
'of highly dynamic cellular response states in epithelial and immune cells associated with '
'specific time points and infection status. We observed that the interferon response in blood '
'preceded the nasopharyngeal response. Moreover, nasopharyngeal immune infiltration occurred '
'early in samples from individuals with only transient infection and later in samples from '
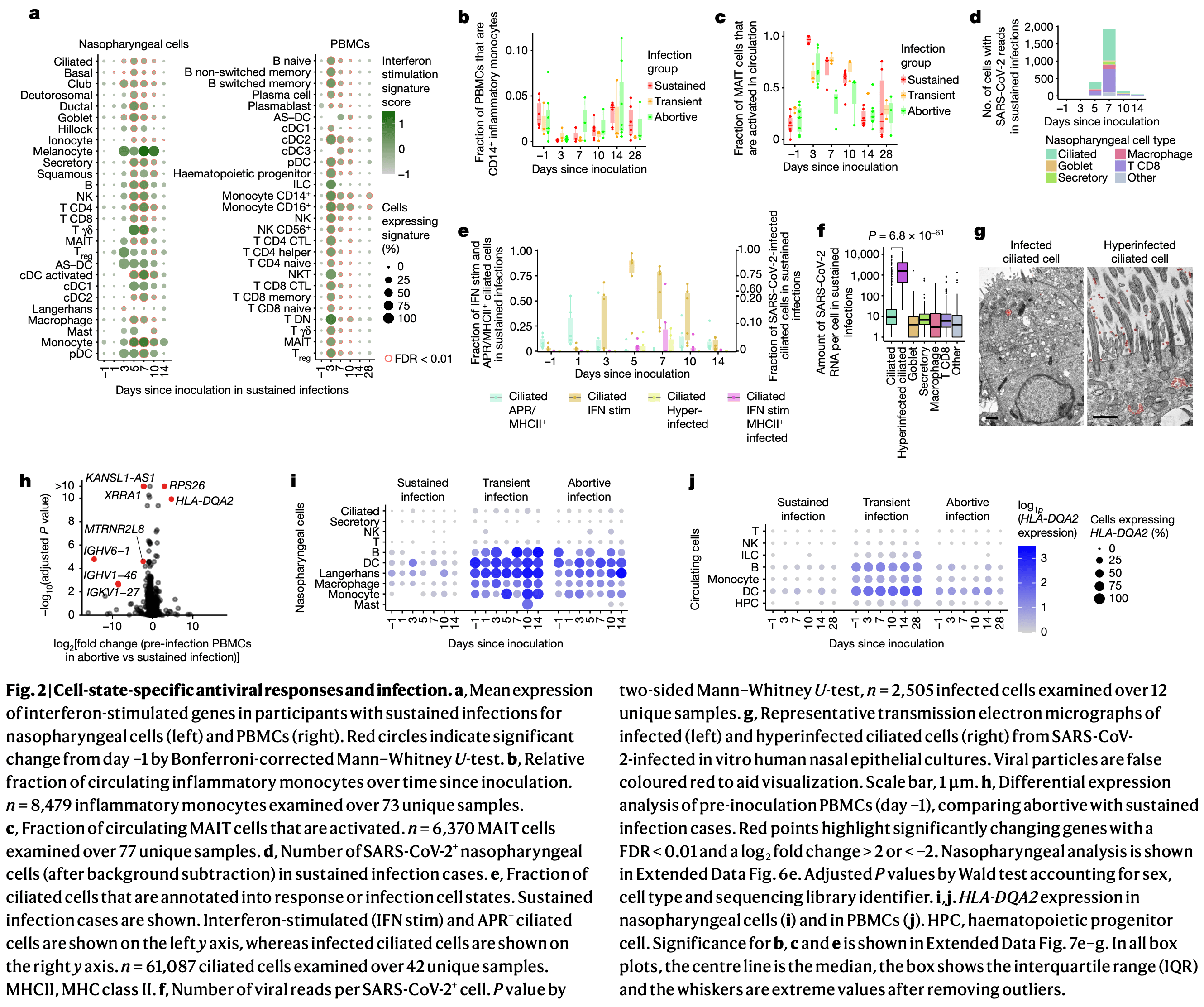
'individuals with sustained infection. High expression of <jats:italic>HLA-DQA2</jats:italic> '
'before inoculation was associated with preventing sustained infection. Ciliated cells showed '
'multiple immune responses and were most permissive for viral replication, whereas '
'nasopharyngeal T\u2009cells and macrophages were infected non-productively. We resolved 54 '
'T\u2009cell states, including acutely activated T\u2009cells that clonally expanded while '
'carrying convergent SARS-CoV-2 motifs. Our new computational pipeline Cell2TCR identifies '
'activated antigen-responding T\u2009cells based on a gene expression signature and clusters '
'these into clonotype groups and motifs. Overall, our detailed time series data can serve as a '
'Rosetta stone for epithelial and immune cell responses and reveals early dynamic responses '
'associated with protection against infection.</jats:p>',
'DOI': '10.1038/s41586-024-07575-x',
'type': 'journal-article',
'created': {'date-parts': [[2024, 6, 19]], 'date-time': '2024-06-19T16:04:49Z', 'timestamp': 1718813089000},
'update-policy': 'http://dx.doi.org/10.1007/springer_crossmark_policy',
'source': 'Crossref',
'is-referenced-by-count': 2,
'title': 'Human SARS-CoV-2 challenge uncovers local and systemic response dynamics',
'prefix': '10.1038',
'author': [ { 'ORCID': 'http://orcid.org/0000-0002-3660-504X',
'authenticated-orcid': False,
'given': 'Rik G. H.',
'family': 'Lindeboom',
'sequence': 'first',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0002-5656-7634',
'authenticated-orcid': False,
'given': 'Kaylee B.',
'family': 'Worlock',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0002-2873-6787',
'authenticated-orcid': False,
'given': 'Lisa M.',
'family': 'Dratva',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0002-3521-5322',
'authenticated-orcid': False,
'given': 'Masahiro',
'family': 'Yoshida',
'sequence': 'additional',
'affiliation': []},
{'given': 'David', 'family': 'Scobie', 'sequence': 'additional', 'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0002-4462-7484',
'authenticated-orcid': False,
'given': 'Helen R.',
'family': 'Wagstaffe',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0002-8075-3816',
'authenticated-orcid': False,
'given': 'Laura',
'family': 'Richardson',
'sequence': 'additional',
'affiliation': []},
{'given': 'Anna', 'family': 'Wilbrey-Clark', 'sequence': 'additional', 'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0001-9938-3176',
'authenticated-orcid': False,
'given': 'Josephine L.',
'family': 'Barnes',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0003-0987-8429',
'authenticated-orcid': False,
'given': 'Lorenz',
'family': 'Kretschmer',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0002-2586-9576',
'authenticated-orcid': False,
'given': 'Krzysztof',
'family': 'Polanski',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0003-4644-0362',
'authenticated-orcid': False,
'given': 'Jessica',
'family': 'Allen-Hyttinen',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0001-9459-9306',
'authenticated-orcid': False,
'given': 'Puja',
'family': 'Mehta',
'sequence': 'additional',
'affiliation': []},
{'given': 'Dinithi', 'family': 'Sumanaweera', 'sequence': 'additional', 'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0001-6813-5217',
'authenticated-orcid': False,
'given': 'Jacqueline M.',
'family': 'Boccacino',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0002-0136-4960',
'authenticated-orcid': False,
'given': 'Waradon',
'family': 'Sungnak',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0001-7366-5466',
'authenticated-orcid': False,
'given': 'Rasa',
'family': 'Elmentaite',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0001-8849-038X',
'authenticated-orcid': False,
'given': 'Ni',
'family': 'Huang',
'sequence': 'additional',
'affiliation': []},
{'given': 'Lira', 'family': 'Mamanova', 'sequence': 'additional', 'affiliation': []},
{ 'ORCID': 'http://orcid.org/0009-0001-3394-0387',
'authenticated-orcid': False,
'given': 'Rakesh',
'family': 'Kapuge',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0001-7293-0774',
'authenticated-orcid': False,
'given': 'Liam',
'family': 'Bolt',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0001-8870-0316',
'authenticated-orcid': False,
'given': 'Elena',
'family': 'Prigmore',
'sequence': 'additional',
'affiliation': []},
{'given': 'Ben', 'family': 'Killingley', 'sequence': 'additional', 'affiliation': []},
{'given': 'Mariya', 'family': 'Kalinova', 'sequence': 'additional', 'affiliation': []},
{'given': 'Maria', 'family': 'Mayer', 'sequence': 'additional', 'affiliation': []},
{'given': 'Alison', 'family': 'Boyers', 'sequence': 'additional', 'affiliation': []},
{'given': 'Alex', 'family': 'Mann', 'sequence': 'additional', 'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0002-0537-6715',
'authenticated-orcid': False,
'given': 'Leo',
'family': 'Swadling',
'sequence': 'additional',
'affiliation': []},
{ 'given': 'Maximillian N. J.',
'family': 'Woodall',
'sequence': 'additional',
'affiliation': []},
{'given': 'Samuel', 'family': 'Ellis', 'sequence': 'additional', 'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0002-8913-0009',
'authenticated-orcid': False,
'given': 'Claire M.',
'family': 'Smith',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0003-0090-861X',
'authenticated-orcid': False,
'given': 'Vitor H.',
'family': 'Teixeira',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0002-6634-5939',
'authenticated-orcid': False,
'given': 'Sam M.',
'family': 'Janes',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0003-1370-9417',
'authenticated-orcid': False,
'given': 'Rachel C.',
'family': 'Chambers',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0002-3927-2084',
'authenticated-orcid': False,
'given': 'Muzlifah',
'family': 'Haniffa',
'sequence': 'additional',
'affiliation': []},
{'given': 'Andrew', 'family': 'Catchpole', 'sequence': 'additional', 'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0003-4573-449X',
'authenticated-orcid': False,
'given': 'Robert',
'family': 'Heyderman',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0002-4774-0853',
'authenticated-orcid': False,
'given': 'Mahdad',
'family': 'Noursadeghi',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0002-7417-3970',
'authenticated-orcid': False,
'given': 'Benny',
'family': 'Chain',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0002-6643-7622',
'authenticated-orcid': False,
'given': 'Andreas',
'family': 'Mayer',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0001-5906-1498',
'authenticated-orcid': False,
'given': 'Kerstin B.',
'family': 'Meyer',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0003-0914-920X',
'authenticated-orcid': False,
'given': 'Christopher',
'family': 'Chiu',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0001-6304-6848',
'authenticated-orcid': False,
'given': 'Marko Z.',
'family': 'Nikolić',
'sequence': 'additional',
'affiliation': []},
{ 'ORCID': 'http://orcid.org/0000-0002-6294-6366',
'authenticated-orcid': False,
'given': 'Sarah A.',
'family': 'Teichmann',
'sequence': 'additional',
'affiliation': []}],
'member': '297',
'published-online': {'date-parts': [[2024, 6, 19]]},
'reference': [ { 'key': '7575_CR1',
'doi-asserted-by': 'publisher',
'DOI': '10.1126/sciimmunol.adj9285',
'volume': '9',
'author': 'HR Wagstaffe',
'year': '2024',
'unstructured': 'Wagstaffe, H. R. et al. Mucosal and systemic immune correlates of viral '
'control after SARS-CoV-2 infection challenge in seronegative adults. '
'Sci. Immunol. 9, eadj9285 (2024).',
'journal-title': 'Sci. Immunol.'},
{ 'key': '7575_CR2',
'doi-asserted-by': 'publisher',
'first-page': '1036',
'DOI': '10.1016/j.cell.2020.04.026',
'volume': '181',
'author': 'D Blanco-Melo',
'year': '2020',
'unstructured': 'Blanco-Melo, D. et al. Imbalanced host response to SARS-CoV-2 drives '
'development of COVID-19. Cell 181, 1036–1045.e9 (2020).',
'journal-title': 'Cell'},
{ 'key': '7575_CR3',
'doi-asserted-by': 'publisher',
'first-page': '718',
'DOI': '10.1126/science.abc6027',
'volume': '369',
'author': 'J Hadjadj',
'year': '2020',
'unstructured': 'Hadjadj, J. et al. Impaired type I interferon activity and inflammatory '
'responses in severe COVID-19 patients. Science 369, 718–724 (2020).',
'journal-title': 'Science'},
{ 'key': '7575_CR4',
'doi-asserted-by': 'publisher',
'first-page': '1419',
'DOI': '10.1016/j.cell.2020.08.001',
'volume': '182',
'author': 'J Schulte-Schrepping',
'year': '2020',
'unstructured': 'Schulte-Schrepping, J. et al. Severe COVID-19 is marked by a '
'dysregulated myeloid cell compartment. Cell 182, 1419–1440.e23 (2020).',
'journal-title': 'Cell'},
{ 'key': '7575_CR5',
'doi-asserted-by': 'publisher',
'first-page': '904',
'DOI': '10.1038/s41591-021-01329-2',
'volume': '27',
'author': 'E Stephenson',
'year': '2021',
'unstructured': 'Stephenson, E. et al. Single-cell multi-omics analysis of the immune '
'response in COVID-19. Nat. Med. 27, 904–916 (2021).',
'journal-title': 'Nat. Med.'},
{ 'key': '7575_CR6',
'doi-asserted-by': 'publisher',
'first-page': '321',
'DOI': '10.1038/s41586-021-04345-x',
'volume': '602',
'author': 'M Yoshida',
'year': '2022',
'unstructured': 'Yoshida, M. et al. Local and systemic responses to SARS-CoV-2 infection '
'in children and adults. Nature 602, 321–327 (2022).',
'journal-title': 'Nature'},
{ 'key': '7575_CR7',
'doi-asserted-by': 'publisher',
'first-page': '1031',
'DOI': '10.1038/s41591-022-01780-9',
'volume': '28',
'author': 'B Killingley',
'year': '2022',
'unstructured': 'Killingley, B. et al. Safety, tolerability and viral kinetics during '
'SARS-CoV-2 human challenge in young adults. Nat. Med. 28, 1031–1041 '
'(2022).',
'journal-title': 'Nat. Med.'},
{ 'key': '7575_CR8',
'doi-asserted-by': 'publisher',
'first-page': '1380',
'DOI': '10.1038/s42003-022-04310-y',
'volume': '5',
'author': 'AC Fears',
'year': '2022',
'unstructured': 'Fears, A. C. et al. The dynamics of γδ T cell responses in nonhuman '
'primates during SARS-CoV-2 infection. Commun. Biol. 5, 1380 (2022).',
'journal-title': 'Commun. Biol.'},
{ 'key': '7575_CR9',
'doi-asserted-by': 'publisher',
'first-page': 'eabq3059',
'DOI': '10.1126/scitranslmed.abq3059',
'volume': '14',
'author': 'JJ Frere',
'year': '2022',
'unstructured': 'Frere, J. J. et al. SARS-CoV-2 infection in hamsters and humans results '
'in lasting and unique systemic perturbations after recovery. Sci. '
'Transl. Med. 14, eabq3059 (2022).',
'journal-title': 'Sci. Transl. Med.'},
{ 'key': '7575_CR10',
'doi-asserted-by': 'publisher',
'first-page': '319',
'DOI': '10.1038/s41587-021-01037-9',
'volume': '40',
'author': 'J Loske',
'year': '2022',
'unstructured': 'Loske, J. et al. Pre-activated antiviral innate immunity in the upper '
'airways controls early SARS-CoV-2 infection in children. Nat. '
'Biotechnol. 40, 319–324 (2022).',
'journal-title': 'Nat. Biotechnol.'},
{ 'key': '7575_CR11',
'doi-asserted-by': 'publisher',
'first-page': '183',
'DOI': '10.1016/S1473-3099(21)00648-4',
'volume': '22',
'author': 'A Singanayagam',
'year': '2022',
'unstructured': 'Singanayagam, A. et al. Community transmission and viral load kinetics '
'of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and '
'unvaccinated individuals in the UK: a prospective, longitudinal, cohort '
'study. Lancet Infect. Dis. 22, 183–195 (2022).',
'journal-title': 'Lancet Infect. Dis.'},
{ 'key': '7575_CR12',
'doi-asserted-by': 'publisher',
'unstructured': 'Rosenheim, J. et al. SARS-CoV-2 human challenge reveals single-gene '
'blood transcriptional biomarkers that discriminate early and late phases '
'of acute respiratory viral infections. Preprint at medRxiv '
'https://doi.org/10.1101/2023.06.01.23290819 (2023).',
'DOI': '10.1101/2023.06.01.23290819'},
{ 'key': '7575_CR13',
'doi-asserted-by': 'publisher',
'unstructured': 'Hinks, T. S. C. & Zhang, X.-W. MAIT cell activation and functions. '
'Front. Immunol. https://doi.org/10.3389/fimmu.2020.01014 (2020).',
'DOI': '10.3389/fimmu.2020.01014'},
{ 'key': '7575_CR14',
'doi-asserted-by': 'publisher',
'DOI': '10.1016/j.xcrm.2022.100557',
'volume': '3',
'author': 'A Chandran',
'year': '2022',
'unstructured': 'Chandran, A. et al. Rapid synchronous type 1 IFN and virus-specific T '
'cell responses characterize first wave non-severe SARS-CoV-2 infections. '
'Cell Rep. Med. 3, 100557 (2022).',
'journal-title': 'Cell Rep. Med.'},
{ 'key': '7575_CR15',
'doi-asserted-by': 'publisher',
'first-page': '283',
'DOI': '10.1038/s41392-020-00426-x',
'volume': '5',
'author': 'K Wang',
'year': '2020',
'unstructured': 'Wang, K. et al. CD147-spike protein is a novel route for SARS-CoV-2 '
'infection to host cells. Signal Transduct. Target. Ther. 5, 283 (2020).',
'journal-title': 'Signal Transduct. Target. Ther.'},
{ 'key': '7575_CR16',
'doi-asserted-by': 'publisher',
'unstructured': 'Brunetti, N. S. et al. SARS-CoV-2 uses CD4 to infect T helper '
'lymphocytes. eLife https://doi.org/10.7554/eLife.84790 (2023).',
'DOI': '10.7554/eLife.84790'},
{ 'key': '7575_CR17',
'doi-asserted-by': 'publisher',
'first-page': '83',
'DOI': '10.1038/s41392-022-00919-x',
'volume': '7',
'author': 'X-R Shen',
'year': '2022',
'unstructured': 'Shen, X.-R. et al. ACE2-independent infection of T lymphocytes by '
'SARS-CoV-2. Signal Transduct. Target. Ther. 7, 83 (2022).',
'journal-title': 'Signal Transduct. Target. Ther.'},
{ 'key': '7575_CR18',
'doi-asserted-by': 'publisher',
'first-page': '2144',
'DOI': '10.3389/fimmu.2018.02144',
'volume': '9',
'author': 'JE Wosen',
'year': '2018',
'unstructured': 'Wosen, J. E., Mukhopadhyay, D., Macaubas, C. & Mellins, E. D. Epithelial '
'MHC class II expression and its role in antigen presentation in the '
'gastrointestinal and respiratory tracts. Front. Immunol. 9, 2144 (2018).',
'journal-title': 'Front. Immunol.'},
{ 'key': '7575_CR19',
'doi-asserted-by': 'publisher',
'first-page': '66',
'DOI': '10.1038/s41588-022-01243-4',
'volume': '55',
'author': 'E Madissoon',
'year': '2023',
'unstructured': 'Madissoon, E. et al. A spatially resolved atlas of the human lung '
'characterizes a gland-associated immune niche. Nat. Genet. 55, 66–77 '
'(2023).',
'journal-title': 'Nat. Genet.'},
{ 'key': '7575_CR20',
'doi-asserted-by': 'publisher',
'first-page': '5834',
'DOI': '10.1038/s41467-021-26045-w',
'volume': '12',
'author': 'AT Shenoy',
'year': '2021',
'unstructured': 'Shenoy, A. T. et al. Antigen presentation by lung epithelial cells '
'directs CD4+ TRM cell function and regulates barrier immunity. Nat. '
'Commun. 12, 5834 (2021).',
'journal-title': 'Nat. Commun.'},
{ 'key': '7575_CR21',
'doi-asserted-by': 'publisher',
'first-page': '2116',
'DOI': '10.4049/jimmunol.158.5.2116',
'volume': '158',
'author': 'GB Rudy',
'year': '1997',
'unstructured': 'Rudy, G. B. & Lew, A. M. The nonpolymorphic MHC class II isotype, '
'HLA-DQA2, is expressed on the surface of B lymphoblastoid cells. J. '
'Immunol. 158, 2116–2125 (1997).',
'journal-title': 'J. Immunol.'},
{ 'key': '7575_CR22',
'doi-asserted-by': 'publisher',
'first-page': '3903',
'DOI': '10.4049/jimmunol.1103048',
'volume': '188',
'author': 'C Lenormand',
'year': '2012',
'unstructured': 'Lenormand, C. et al. HLA-DQA2 and HLA-DQB2 genes are specifically '
'expressed in human Langerhans cells and encode a new HLA class II '
'molecule. J. Immunol. 188, 3903–3911 (2012).',
'journal-title': 'J. Immunol.'},
{ 'key': '7575_CR23',
'doi-asserted-by': 'publisher',
'first-page': '538',
'DOI': '10.1038/s41556-021-00681-2',
'volume': '23',
'author': 'M Saichi',
'year': '2021',
'unstructured': 'Saichi, M. et al. Single-cell RNA sequencing of blood antigen-presenting '
'cells in severe COVID-19 reveals multi-process defects in antiviral '
'immunity. Nat. Cell Biol. 23, 538–551 (2021).',
'journal-title': 'Nat. Cell Biol.'},
{ 'key': '7575_CR24',
'doi-asserted-by': 'crossref',
'unstructured': 'Policard, M., Jain, S., Rego, S. & Dakshanamurthy S. Immune '
'characterization and profiles of SARS-CoV-2 infected patients reveals '
'potential host therapeutic targets and SARS-CoV-2 oncogenesis mechanism. '
'Virus Res. 301, 198464 (2021).',
'DOI': '10.1016/j.virusres.2021.198464'},
{ 'key': '7575_CR25',
'doi-asserted-by': 'publisher',
'unstructured': 'Kaneko, N. et al. Temporal changes in T cell subsets and expansion of '
'cytotoxic CD4 T cells in the lungs in severe COVID-19. Clin. Immunol. '
'https://doi.org/10.1016/j.clim.2022.108991 (2022).',
'DOI': '10.1016/j.clim.2022.108991'},
{ 'key': '7575_CR26',
'doi-asserted-by': 'publisher',
'first-page': '1340',
'DOI': '10.1016/j.cell.2020.10.001',
'volume': '183',
'author': 'BJ Meckiff',
'year': '2020',
'unstructured': 'Meckiff, B. J. et al. Imbalance of regulatory and cytotoxic '
'SARS-CoV-2-reactive CD4 T cells in COVID-19. Cell 183, 1340–1353.e16 '
'(2020).',
'journal-title': 'Cell'},
{ 'key': '7575_CR27',
'doi-asserted-by': 'publisher',
'first-page': '274',
'DOI': '10.1038/nm.2612',
'volume': '18',
'author': 'TM Wilkinson',
'year': '2012',
'unstructured': 'Wilkinson, T. M. et al. Preexisting influenza-specific CD4+ T cells '
'correlate with disease protection against influenza challenge in humans. '
'Nat. Med. 18, 274–280 (2012).',
'journal-title': 'Nat. Med.'},
{ 'key': '7575_CR28',
'doi-asserted-by': 'publisher',
'first-page': '983',
'DOI': '10.4049/jimmunol.1600318',
'volume': '197',
'author': 'JM Dan',
'year': '2016',
'unstructured': 'Dan, J. M. et al. A cytokine-independent approach to identify '
'antigen-specific human germinal center T follicular helper cells and '
'rare antigen-specific CD4+ T cells in blood. J. Immunol. 197, 983–993 '
'(2016).',
'journal-title': 'J. Immunol.'},
{ 'key': '7575_CR29',
'doi-asserted-by': 'publisher',
'first-page': '94',
'DOI': '10.1126/science.274.5284.94',
'volume': '274',
'author': 'JD Altman',
'year': '1996',
'unstructured': 'Altman, J. D. et al. Phenotypic analysis of antigen-specific T '
'lymphocytes. Science 274, 94–96 (1996).',
'journal-title': 'Science'},
{ 'key': '7575_CR30',
'doi-asserted-by': 'publisher',
'first-page': '772',
'DOI': '10.4049/jimmunol.2200154',
'volume': '209',
'author': 'G Wigerblad',
'year': '2022',
'unstructured': 'Wigerblad, G. et al. Single-cell analysis reveals the range of '
'transcriptional states of circulating human neutrophils. J. Immunol. '
'209, 772–782 (2022).',
'journal-title': 'J. Immunol.'},
{ 'key': '7575_CR31',
'unstructured': 'Ren, X. et al. COVID-19 immune features revealed by a large-scale '
'single-cell transcriptome atlas. Cell 184, 1895–1913.e19 (2021).'},
{ 'key': '7575_CR32',
'doi-asserted-by': 'crossref',
'unstructured': 'Liu, C. et al. Time-resolved systems immunology reveals a late juncture '
'linked to fatal COVID-19. Cell 184, 1836–1857.e22 (2021).',
'DOI': '10.1016/j.cell.2021.02.018'},
{ 'key': '7575_CR33',
'unstructured': 'COvid-19 Multi-omics Blood ATlas (COMBAT) Consortium. A blood atlas of '
'COVID-19 defines hallmarks of disease severity and specificity. Cell '
'185, 916–938.e58 (2022).'},
{ 'key': '7575_CR34',
'unstructured': 'ISARIC 4C. Coronavirus Clinical Characterisation Consortium. Site set-up '
'https://isaric4c.net/protocols (2020).'},
{ 'key': '7575_CR35',
'doi-asserted-by': 'publisher',
'unstructured': 'Tang, Y. et al. Human nasopharyngeal swab processing for viable '
'single-cell suspension v1. protocols.io '
'https://doi.org/10.17504/protocols.io.bjhkkj4w (2020).',
'DOI': '10.17504/protocols.io.bjhkkj4w'},
{ 'key': '7575_CR36',
'doi-asserted-by': 'publisher',
'first-page': '4713',
'DOI': '10.1016/j.cell.2021.07.023',
'volume': '184',
'author': 'CGK Ziegler',
'year': '2021',
'unstructured': 'Ziegler, C. G. K. et al. Impaired local intrinsic immunity to SARS-CoV-2 '
'infection in severe COVID-19. Cell 184, 4713–4733.e22 (2021).',
'journal-title': 'Cell'},
{ 'key': '7575_CR37',
'doi-asserted-by': 'publisher',
'first-page': 'eabo0510',
'DOI': '10.1126/science.abo0510',
'volume': '376',
'author': 'C Suo',
'year': '2022',
'unstructured': 'Suo, C. et al. Mapping the developing human immune system across organs. '
'Science 376, eabo0510 (2022).',
'journal-title': 'Science'},
{ 'key': '7575_CR38',
'doi-asserted-by': 'publisher',
'DOI': '10.1093/gigascience/giaa151',
'volume': '9',
'author': 'MD Young',
'year': '2020',
'unstructured': 'Young, M. D. & Behjati, S. SoupX removes ambient RNA contamination from '
'droplet-based single-cell RNA sequencing data. Gigascience 9, giaa151 '
'(2020).',
'journal-title': 'Gigascience'},
{ 'key': '7575_CR39',
'doi-asserted-by': 'publisher',
'first-page': '615',
'DOI': '10.1038/s41592-020-0820-1',
'volume': '17',
'author': 'H Heaton',
'year': '2020',
'unstructured': 'Heaton, H. et al. Souporcell: robust clustering of single-cell RNA-seq '
'data by genotype without reference genotypes. Nat. Methods 17, 615–620 '
'(2020).',
'journal-title': 'Nat. Methods'},
{ 'key': '7575_CR40',
'doi-asserted-by': 'publisher',
'first-page': '1289',
'DOI': '10.1038/s41592-019-0619-0',
'volume': '16',
'author': 'I Korsunsky',
'year': '2019',
'unstructured': 'Korsunsky, I. et al. Fast, sensitive and accurate integration of '
'single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).',
'journal-title': 'Nat. Methods'},
{ 'key': '7575_CR41',
'doi-asserted-by': 'publisher',
'DOI': '10.1038/s41598-019-41695-z',
'volume': '9',
'author': 'VA Traag',
'year': '2019',
'unstructured': 'Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: '
'guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).',
'journal-title': 'Sci. Rep.'},
{ 'key': '7575_CR42',
'doi-asserted-by': 'publisher',
'first-page': 'eabl5197',
'DOI': '10.1126/science.abl5197',
'volume': '376',
'author': 'C Domínguez Conde',
'year': '2022',
'unstructured': 'Domínguez Conde, C. et al. Cross-tissue immune cell analysis reveals '
'tissue-specific features in humans. Science 376, eabl5197 (2022).',
'journal-title': 'Science'},
{ 'key': '7575_CR43',
'doi-asserted-by': 'publisher',
'first-page': '4817',
'DOI': '10.1093/bioinformatics/btaa611',
'volume': '36',
'author': 'G Sturm',
'year': '2020',
'unstructured': 'Sturm, G. et al. Scirpy: a Scanpy extension for analyzing single-cell '
'T-cell receptor-sequencing data. Bioinformatics 36, 4817–4818 (2020).',
'journal-title': 'Bioinformatics'},
{ 'key': '7575_CR44',
'doi-asserted-by': 'publisher',
'first-page': '40',
'DOI': '10.1038/s41587-023-01734-7',
'volume': '42',
'author': 'C Suo',
'year': '2023',
'unstructured': 'Suo, C. et al. Dandelion uses the single-cell adaptive immune receptor '
'repertoire to explore lymphocyte developmental origins. Nat. Biotechnol. '
'42, 40–51 (2023).',
'journal-title': 'Nat. Biotechnol.'},
{ 'key': '7575_CR45',
'doi-asserted-by': 'publisher',
'DOI': '10.1186/s13059-014-0550-8',
'volume': '15',
'author': 'MI Love',
'year': '2014',
'unstructured': 'Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change '
'and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 '
'(2014).',
'journal-title': 'Genome Biol.'},
{ 'key': '7575_CR46',
'doi-asserted-by': 'publisher',
'DOI': '10.1186/s13059-017-1382-0',
'volume': '19',
'author': 'FA Wolf',
'year': '2018',
'unstructured': 'Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell '
'gene expression data analysis. Genome Biol. 19, 15 (2018).',
'journal-title': 'Genome Biol.'},
{ 'key': '7575_CR47',
'doi-asserted-by': 'publisher',
'first-page': 'e8746',
'DOI': '10.15252/msb.20188746',
'volume': '15',
'author': 'MD Luecken',
'year': '2019',
'unstructured': 'Luecken, M. D. & Theis, F. J. Current best practices in single-cell '
'RNA-seq analysis: a tutorial. Mol. Syst. Biol. 15, e8746 (2019).',
'journal-title': 'Mol. Syst. Biol.'},
{ 'key': '7575_CR48',
'doi-asserted-by': 'publisher',
'unstructured': 'Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep '
'generative modeling for single-cell transcriptomics. Nat. Methods '
'https://doi.org/10.1038/s41592-018-0229-2 (2018).',
'DOI': '10.1038/s41592-018-0229-2'},
{ 'key': '7575_CR49',
'doi-asserted-by': 'publisher',
'first-page': 'e68605',
'DOI': '10.7554/eLife.68605',
'volume': '10',
'author': 'K Mayer-Blackwell',
'year': '2021',
'unstructured': 'Mayer-Blackwell, K. et al. TCR meta-clonotypes for biomarker discovery '
'with enabled identification of public, HLA-restricted clusters of '
'SARS-CoV-2 TCRs. eLife 10, e68605 (2021).',
'journal-title': 'eLife'},
{ 'key': '7575_CR50',
'doi-asserted-by': 'publisher',
'unstructured': 'Csárdi, G. et al. Igraph. Zenodo https://doi.org/10.5281/ZENODO.3630268 '
'(2023).',
'DOI': '10.5281/ZENODO.3630268'},
{ 'key': '7575_CR51',
'doi-asserted-by': 'publisher',
'first-page': '3645',
'DOI': '10.1093/bioinformatics/btx469',
'volume': '33',
'author': 'O Wagih',
'year': '2017',
'unstructured': 'Wagih, O. ggseqlogo: a versatile R package for drawing sequence logos. '
'Bioinformatics 33, 3645–3647 (2017).',
'journal-title': 'Bioinformatics'},
{ 'key': '7575_CR52',
'doi-asserted-by': 'publisher',
'first-page': '2272',
'DOI': '10.1093/bioinformatics/btz921',
'volume': '36',
'author': 'A Tareen',
'year': '2020',
'unstructured': 'Tareen, A. & Kinney, J. B. Logomaker: beautiful sequence logos in '
'Python. Bioinformatics 36, 2272–2274 (2020).',
'journal-title': 'Bioinformatics'},
{ 'key': '7575_CR53',
'first-page': '844',
'volume': '9',
'author': 'M Titsias',
'year': '2010',
'unstructured': 'Titsias, M. & Lawrence, N. D. Bayesian Gaussian process latent variable '
'model. Proc. Mach. Learn. Res. 9, 844–851 (2010).',
'journal-title': 'Proc. Mach. Learn. Res.'},
{ 'key': '7575_CR54',
'unstructured': 'Bingham, E. et al. Pyro: deep universal probabilistic programming. J. '
'Mach. Learn. Res. 20, 1–6 (2019)'},
{ 'key': '7575_CR55',
'unstructured': 'Bonilla, E. V., Chai, K. & Williams, C. Multi-task Gaussian process '
'prediction. In Proc. Advances in Neural Information Processing Systems '
'(eds Platt, J. et al.) (Curran Associates, 2007).'},
{ 'key': '7575_CR56',
'unstructured': 'Gardner, J. R., Pleiss, G., Weinberger, K. Q., Bindel, D. & Wilson, A. '
'G. GPyTorch: blackbox matrix-matrix gaussian process inference with GPU '
'acceleration. in Advances in Neural Information Processing Systems 31 '
'(eds Samy Bengio, S. et al.) 7587–7597 (NeurIPS, 2018).'},
{ 'key': '7575_CR57',
'doi-asserted-by': 'publisher',
'unstructured': 'Uddin, I. et al. An economical, quantitative, and robust protocol for '
'high-throughput T cell receptor sequencing from tumor or blood. Methods '
'Mol. Biol. https://doi.org/10.1007/978-1-4939-8885-3_2 (2019).',
'DOI': '10.1007/978-1-4939-8885-3_2'},
{ 'key': '7575_CR58',
'doi-asserted-by': 'publisher',
'first-page': '1267',
'DOI': '10.3389/fimmu.2017.01267',
'volume': '8',
'author': 'T Oakes',
'year': '2017',
'unstructured': 'Oakes, T. et al. Quantitative characterization of the T cell receptor '
'repertoire of naïve and memory subsets using an integrated experimental '
'and computational pipeline which is robust, economical, and versatile. '
'Front. Immunol. 8, 1267 (2017).',
'journal-title': 'Front. Immunol.'},
{ 'key': '7575_CR59',
'doi-asserted-by': 'publisher',
'first-page': '876',
'DOI': '10.1093/bioinformatics/btaa758',
'volume': '37',
'author': 'T Peacock',
'year': '2021',
'unstructured': 'Peacock, T., Heather, J. M., Ronel, T. & Chain, B. Decombinator V4: an '
'improved AIRR compliant-software package for T-cell receptor sequence '
'annotation? Bioinformatics 37, 876–878 (2021).',
'journal-title': 'Bioinformatics'},
{ 'key': '7575_CR60',
'doi-asserted-by': 'publisher',
'first-page': 'e2213264120',
'DOI': '10.1073/pnas.2213264120',
'volume': '120',
'author': 'A Mayer',
'year': '2023',
'unstructured': 'Mayer, A. & Callan, C. G. Jr Measures of epitope binding degeneracy from '
'T cell receptor repertoires. Proc. Natl Acad. Sci. USA 120, e2213264120 '
'(2023).',
'journal-title': 'Proc. Natl Acad. Sci. USA'},
{ 'key': '7575_CR61',
'doi-asserted-by': 'publisher',
'first-page': '179',
'DOI': '10.12688/wellcomeopenres.16051.2',
'volume': '5',
'author': 'JB Augusto',
'year': '2020',
'unstructured': 'Augusto, J. B. et al. Healthcare Workers Bioresource: study outline and '
'baseline characteristics of a prospective healthcare worker cohort to '
'study immune protection and pathogenesis in COVID-19. Wellcome Open Res. '
'5, 179 (2020).',
'journal-title': 'Wellcome Open Res.'},
{ 'key': '7575_CR62',
'doi-asserted-by': 'publisher',
'first-page': '1293',
'DOI': '10.1038/s41564-024-01658-1',
'volume': '9',
'author': 'MNJ Woodall',
'year': '2024',
'unstructured': 'Woodall, M. N. J. et al. Age-specific nasal epithelial responses to '
'SARS-CoV-2 infection. Nat. Microbiol. 9, 1293–1311 (2024).',
'journal-title': 'Nat. Microbiol.'},
{ 'key': '7575_CR63',
'doi-asserted-by': 'publisher',
'first-page': '4255',
'DOI': '10.1113/JP281499',
'volume': '599',
'author': 'MNJ Woodall',
'year': '2021',
'unstructured': 'Woodall, M. N. J., Masonou, T., Case, K.-M. & Smith, C. M. Human models '
'for COVID-19 research. J. Physiol. 599, 4255–4267 (2021).',
'journal-title': 'J. Physiol.'}],
'container-title': 'Nature',
'original-title': [],
'language': 'en',
'link': [ { 'URL': 'https://www.nature.com/articles/s41586-024-07575-x.pdf',
'content-type': 'application/pdf',
'content-version': 'vor',
'intended-application': 'text-mining'},
{ 'URL': 'https://www.nature.com/articles/s41586-024-07575-x',
'content-type': 'text/html',
'content-version': 'vor',
'intended-application': 'text-mining'},
{ 'URL': 'https://www.nature.com/articles/s41586-024-07575-x.pdf',
'content-type': 'application/pdf',
'content-version': 'vor',
'intended-application': 'similarity-checking'}],
'deposited': { 'date-parts': [[2024, 6, 19]],
'date-time': '2024-06-19T16:08:02Z',
'timestamp': 1718813282000},
'score': 1,
'resource': {'primary': {'URL': 'https://www.nature.com/articles/s41586-024-07575-x'}},
'subtitle': [],
'short-title': [],
'issued': {'date-parts': [[2024, 6, 19]]},
'references-count': 63,
'alternative-id': ['7575'],
'URL': 'http://dx.doi.org/10.1038/s41586-024-07575-x',
'relation': {},
'ISSN': ['0028-0836', '1476-4687'],
'subject': [],
'container-title-short': 'Nature',
'published': {'date-parts': [[2024, 6, 19]]},
'assertion': [ { 'value': '24 February 2023',
'order': 1,
'name': 'received',
'label': 'Received',
'group': {'name': 'ArticleHistory', 'label': 'Article History'}},
{ 'value': '16 May 2024',
'order': 2,
'name': 'accepted',
'label': 'Accepted',
'group': {'name': 'ArticleHistory', 'label': 'Article History'}},
{ 'value': '19 June 2024',
'order': 3,
'name': 'first_online',
'label': 'First Online',
'group': {'name': 'ArticleHistory', 'label': 'Article History'}},
{ 'value': 'R.G.H.L., L.M.D., R.E. and S.A.T. are inventors on a filed patent that is '
'related to the detection and application of activated T\u2009cells. In the past '
'3\u2009years, S.A.T. has received remuneration for scientific advisory board '
'membership from Sanofi, GlaxoSmithKline, Foresite Labs and Qiagen. S.A.T. is a '
'co-founder and holds equity in Transition Bio and Ensocell. From 8 January '
'2024, S.A.T. is a part-time employee of GlaxoSmithKline. R.E. is a co-founder '
'and equity holder in Ensocell. P.M. is a Medical Research Council '
'(MRC)-GlaxoSmithKline EMINENT clinical training fellow with project funding '
'unrelated to the topic of this work and receives co-funding from the National '
'Institute for Health Research (NIHR) University College London Hospitals (UCLH) '
'Biomedical Research Centre. P.M. reports consultancy fees from SOBI, AbbVie, '
'UCB, Lilly, Boehringer Ingelheim and EUSA Pharma, all unrelated to this study. '
'S.M.J. has received fees for advisory board membership in the last three years '
'from Bard1 Lifescience. He has received grant income from GRAIL Inc. He is an '
'unpaid member of a GRAIL advisory board. He has received lecture fees for '
'academic meetings from Cheisi and AstraZeneca. His wife works for AstraZeneca. '
'R.C.C. has research collaborations with Chiesi Chiesi Farmaceutici S.p.A. and '
'GSK and receives consulting fees from Vicore, outside the submitted work.\xa0'
'A.\xa0Mann, A.C., M.K., M.M. and A.B. are full-time employees at hVIVO '
'Services. The other authors declare no competing interests.',
'order': 1,
'name': 'Ethics',
'group': {'name': 'EthicsHeading', 'label': 'Competing interests'}}]}