Crowdsourcing Temporal Transcriptomic Coronavirus Host Infection Data: resources, guide, and novel insights
Cov Infection James Flynn, Mehdi M Ahmadi, Chase T Mcfarland, Michael D Kubal, Mark A Taylor, Zhang Cheng, Enrique C Torchia, Michael G Edwards
doi:10.1093/biomethods/bpad033/7420215
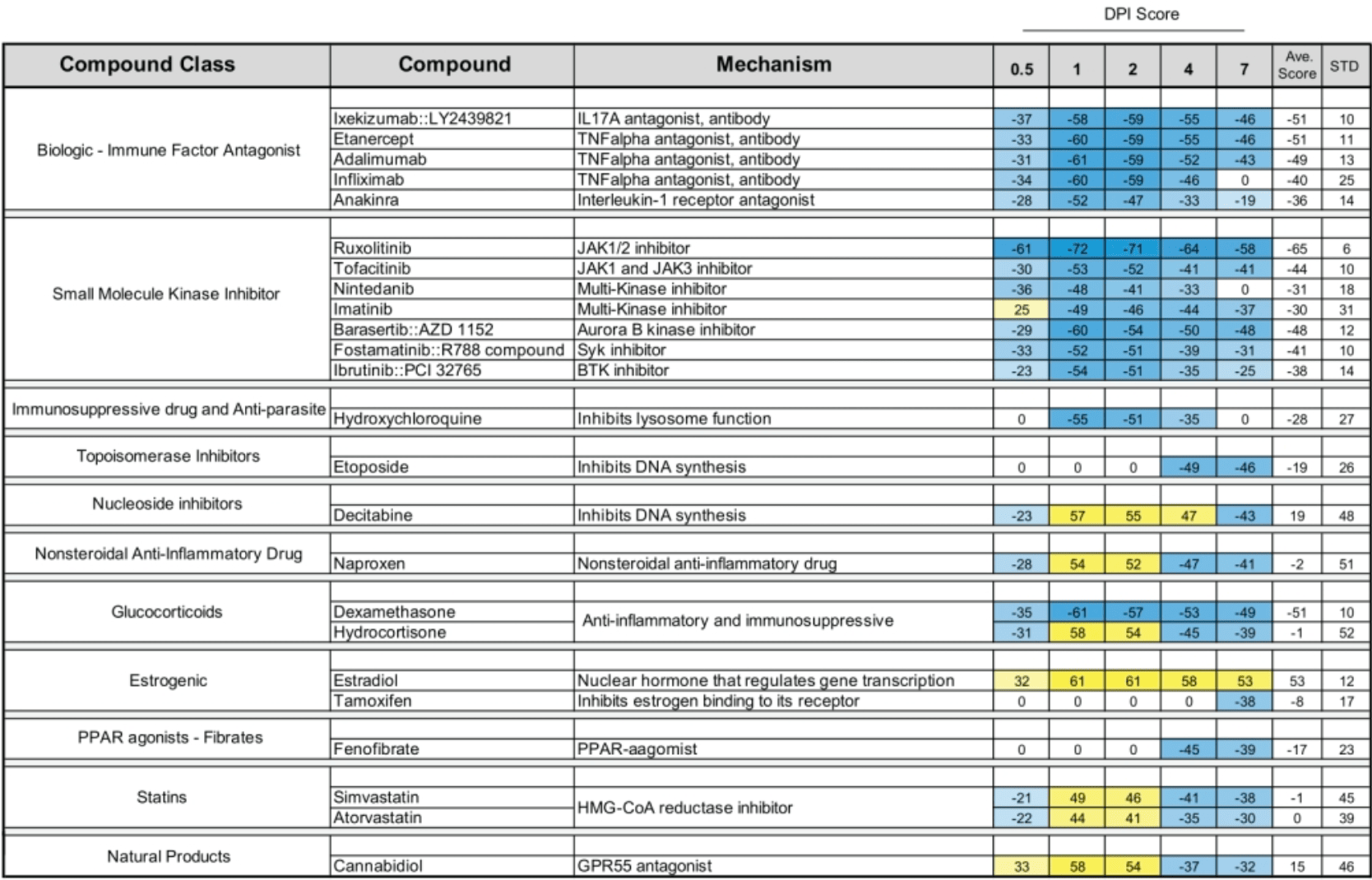
The emergence of SARS-CoV-2 reawakened the need to rapidly understand the molecular etiologies, pandemic potential, and prospective treatments of infectious agents. The lack of existing data on SARS-CoV-2 hampered early attempts to treat severe forms of COVID-19 during the pandemic. This study coupled existing transcriptomic data from SARS-CoV-1 lung infection animal studies with crowdsourcing statistical approaches to derive temporal meta-signatures of host responses during early viral accumulation and subsequent clearance stages. Unsupervised and supervised machine learning approaches identified top dysregulated genes and potential biomarkers (e.g., CXCL10, BEX2, and ADM). Temporal meta-signatures revealed distinct gene expression programs with biological implications to a series of host responses underlying sustained Cxcl10 expression and Stat signaling. Cell cycle switched from G1/G0 phase genes, early in infection, to a G2/M gene signature during late infection that correlated with the enrichment of DNA Damage Response and Repair genes. The SARS-CoV-1 meta-signatures were shown to closely emulate human SARS-CoV-2 host responses from emerging RNAseq, single cell and proteomics data with early monocyte-macrophage activation followed by lymphocyte proliferation. The circulatory hormone adrenomedullin was observed as maximally elevated in elderly patients that died from COVID-19. Stage-specific correlations to compounds with potential to treat COVID-19 and future coronavirus infections were in part validated by a subset of twenty-four that are in clinical trials to treat COVID-19. This study represents a roadmap to leverage existing data in the public domain to derive novel molecular and biological insights and potential treatments to emerging human pathogens.
Competing Interests JF, MK and ZC work for Illumina, the commercial developer of BSCE. ME, ET, MT, CM, MA report no competing interests.
Author contributions JF, ME and ET were responsible for the conception and design of the studies, performed bioinformatics analyses, biological interpretation and manuscript drafting. MK, ZC, MT, CM and MA performed bioinformatics analyses and created the interactive portal. All authors had access to and assisted in data interpretation. All authors helped to critically revise the intellectual content of the manuscript and approved the final submission.
References
Abdelrahman, Li, Wang, Comparative Review of SARS-CoV-2, SARS-CoV, MERS-CoV, and Influenza A Respiratory Viruses, Front Immunol
Agostini, Calabrese, Rea, Facco, Tosoni et al., Cxcr3 and its ligand CXCL10 are expressed by inflammatory cells infiltrating lung allografts and mediate chemotaxis of T cells at sites of rejection, The American journal of pathology
Alvarez, Zhou, Witta, Freed, Characterization of the Bex gene family in humans, mice, and rats, Gene
Andargie, Tsuji, Seifuddin, Jang, Yuen et al., Cell-free DNA maps COVID-19 tissue injury and risk of death and can cause tissue injury, JCI Insight
Asada, Hara, Marutsuka, Kitamura, Tsuji et al., Novel distribution of adrenomedullin-immunoreactive cells in human tissues, Histochem Cell Biol
Ashburner, Ball, Blake, Botstein, Butler et al., Gene ontology: tool for the unification of biology. The Gene Ontology Consortium, Nat Genet
Athar, Füllgrabe, George, Iqbal, Huerta et al., ArrayExpress update -from bulk to single-cell expression data, Nucleic Acids Research
Aucamp, Bronkhorst, Badenhorst, Pretorius, The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature, Biological Reviews
Bahadar, Ullah, Adlat, Sah, Zaw Myint et al., Analyzing differentially expressed genes and pathways of Bex2-deficient mouse lung via RNA-Seq, Turk J Biol
Bao, Deng, Huang, Gao, Liu et al., The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice, Nature
Barretina, Caponigro, Stransky, Venkatesan, Margolin et al., The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity, Nature
Barrett, Wilhite, Ledoux, Evangelista, Kim et al., NCBI GEO: archive for functional genomics data sets-update, Nucleic Acids Research
Behjati, Huch, Van Boxtel, Karthaus, Wedge et al., Genome sequencing of normal cells reveals developmental lineages and mutational processes, Nature
Bernal, Andrews, Gower, Gallagher, Simmons et al., Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant, New England Journal of Medicine
Blet, Deniau, Geven, Sadoune, Caillard et al., Adrecizumab, a non-neutralizing anti-adrenomedullin antibody, improves haemodynamics and attenuates myocardial oxidative stress in septic rats, Intensive Care Med Exp
Borton, Rashid, Dreier, Taylor, Multiple Levels of Regulation of Sororin by Cdk1 and Aurora B, Journal of Cellular Biochemistry
Boutelle, Attardi, p53 and Tumor Suppression: It Takes a Network, Trends in Cell Biology
Brady, Boggan, Bowie, Neill, Schlafen-1 Causes a Cell Cycle Arrest by Inhibiting Induction of Cyclin D1 *, Journal of Biological Chemistry
Branzei, Foiani, Regulation of DNA repair throughout the cell cycle, Nature Reviews Molecular Cell Biology
Brugarolas, Chandrasekaran, Gordon, Beach, Jacks et al., Radiation-induced cell cycle arrest compromised by p21 deficiency, Nature
Callahan, Hawks, Crawford, Lehman, Morrison et al., The Pro-Inflammatory Chemokines CXCL9, CXCL10 and CXCL11 Are Upregulated Following SARS-CoV-2 Infection in an AKT-Dependent Manner, Viruses
Castelli, Cimini, Ferri, Cytokine Storm in COVID-19: "When You Come Out of the Storm, You Won't Be the Same Person Who Walked in, Front Immunol
Chandrasekaran, Idelchik, Melendez, Redox control of senescence and age-related disease, Redox Biol
Chang, Lin, Sibille, Tseng, Meta-analysis methods for combining multiple expression profiles: comparisons, statistical characterization and an application guideline, BMC Bioinformatics
Channappanavar, Anthony, Vijay, Mack, Zhao et al., Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice, Cell Host Microbe
Charlton, Babady, Ginocchio, Hatchette, Jerris et al., Practical Guidance for Clinical Microbiology Laboratories: Viruses Causing Acute Respiratory Tract Infections, Clinical Microbiology Reviews
Cheeseman, The kinetochore, Cold Spring Harb Perspect Biol
Cook, Brown, Alexander, March, Morgan et al., Lessons learned from the fate of AstraZeneca's drug pipeline: a five-dimensional framework, Nat Rev Drug Discov
Coperchini, Chiovato, Rotondi, Interleukin-6, CXCL10 and infiltrating macrophages in COVID-19-related cytokine storm: not one for all but all for one!, Front Immunol
Costa, Da Luz, Campos, Lopes, Jpdv et al., Can SARS-CoV-2 induce hematologic malignancies in predisposed individuals? A case series and review of the literature, Hematology, Transfusion and Cell Therapy
Deniau, Takagi, Asakage, Mebazaa, Adrecizumab: an investigational agent for the biomarkerguided treatment of sepsis, Expert Opin Investig Drugs
Elkhateeb, Tag-El-Din-Hassan, Sasaki, Torigoe, Morimatsu et al., The role of mouse 2',5'-oligoadenylate synthetase 1 paralogs, Infect Genet Evol
Excler, Saville, Berkley, Kim, Vaccine development for emerging infectious diseases, Nat Med
Fajgenbaum, June, Cytokine storm, New England Journal of Medicine
Fernandez, Torchia, Isolation and Staining of Mouse Skin Keratinocytes for Cell Cycle Specific Analysis of Cellular Protein Expression by Mass Cytometry, J Vis Exp
Filbin, Mehta, Schneider, Kays, Guess et al., Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions, Cell Reports Medicine
Frieman, Chen, Morrison, Whitmore, Funkhouser et al., SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism, PLoS Pathog
Galili, 'callaghan, Sidi, Sievert, heatmaply: an R package for creating interactive cluster heatmaps for online publishing, Bioinformatics
Galton, Vox Populi, Nature
Gar, Thorand, Herder, Sujana, Heier et al., Association of circulating MR-proADM with all-cause and cardiovascular mortality in the general population: Results from the KORA F4 cohort study, PLoS One
Gebhard, Regitz-Zagrosek, Neuhauser, Morgan, Klein, Gene Ontology Resource. The Gene Ontology resource: enriching a GOld mine, Nucleic Acids Res
Gerdes, Lemke, Baisch, Wacker, Schwab et al., Cell cycle analysis of a cell proliferationassociated human nuclear antigen defined by the monoclonal antibody Ki-67, The Journal of Immunology
Glass Gv, Primary, Secondary, and Meta-Analysis of Research, Educational Researcher
Glass, Subbarao, Murphy, Murphy, Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice, J Immunol
Gorbalenya, Baker, Baric, De Groot, Drosten et al., The species Severe acute respiratory syndrome-related coronavirus : classifying 2019-nCoV and naming it SARS-CoV-2, Nat Microbiol
Gordon, Jang, Bouhaddou, Xu, Obernier et al., A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug-Repurposing, Nature
Gregoriano, Koch, Kutz, Haubitz, Conen et al., The vasoactive peptide MRpro-adrenomedullin in COVID-19 patients: an observational study, Clin Chem Lab Med
Groom, Luster, CXCR3 ligands: redundant, collaborative and antagonistic functions, Immunol Cell Biol
Gu, Hübschmann, Make Interactive Complex Heatmaps in R, Bioinformatics
Gutiérrez-Caballero, Cebollero, Pendás, Shugoshins: from protectors of cohesion to versatile adaptors at the centromere, Trends in Genetics
Gwinn, Auerbach, Parham, Stout, Waidyanatha et al., Evaluation of 5-day In Vivo Rat Liver and Kidney With High-throughput Transcriptomics for Estimating Benchmark Doses of Apical Outcomes, Toxicol Sci
Haller, Staeheli, Kochs, Interferon-induced Mx proteins in antiviral host defense, Biochimie
Hamid, Hu, Roslin, Ling, Greenwood et al., Data Integration in Genetics and Genomics: Methods and Challenges, Hum Genomics Proteomics
Hammond, Loghavi, Clonal haematopoiesis of emerging significance, Pathology
Hartl, De Luca, Kostikova, Laramie, Kennedy et al., Translational precision medicine: an industry perspective, J Transl Med
Hayward, Alfonso-Pérez, Gruneberg, Orchestration of the spindle assembly checkpoint by CDK1-cyclin B1, FEBS Lett
Henglein, Chenivesse, Wang, Eick, Bréchot, Structure and cell cycle-regulated transcription of the human cyclin A gene, Proceedings of the National Academy of Sciences
Hoffmann, Kleine-Weber, Schroeder, Krüger, Herrler et al., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell
Holmfeldt, Ganuza, Marathe, He, Hall et al., Functional screen identifies regulators of murine hematopoietic stem cell repopulation, J Exp Med
Hupf, Mustroph, Hanses, Evert, Maier et al., RNA-expression of adrenomedullin is increased in patients with severe COVID-19, Crit Care
Hussell, Goulding, Structured regulation of inflammation during respiratory viral infection, Lancet Infect Dis
Igarashi, Nakatsu, Yamashita, Ono, Ohno et al., Open TG-GATEs: a largescale toxicogenomics database, Nucleic Acids Research
Jacobs, Yeager, Zhou, Wacholder, Wang et al., Detectable clonal mosaicism and its relationship to aging and cancer, Nature Genetics
Jafarzadeh, Gosain, Mortazavi, Nemati, Jafarzadeh et al., Review SARS-CoV-2 Infection: A Possible Risk Factor for Incidence and Recurrence of Cancers, International Journal of Hematology-Oncology and Stem Cell Research
Jin, Bai, He, Wu, Liu et al., Gender Differences in Patients With COVID-19: Focus on Severity and Mortality, Front Public Health
Kaeffer, Zeder-Lutz, Simonin, Lecat, GPRASP/ARMCX Protein Family: Potential Involvement in Health and Diseases Revealed by their Novel Interacting Partners, Curr Top Med Chem
Karita, Dong, Johnston, Neuzil, Paasche-Orlow et al., Trajectory of Viral RNA Load Among Persons With Incident SARS-CoV-2 G614 Infection (Wuhan Strain) in Association With COVID-19 Symptom Onset and Severity, JAMA Network Open
Kazi, Kabir, Rönnstrand, Brain-Expressed X-linked (BEX) proteins in human cancers, Biochim Biophys Acta
Khomich, Kochetkov, Bartosch, Ivanov, Redox Biology of Respiratory Viral Infections, Viruses
Kitamura, Kangawa, Kawamoto, Ichiki, Nakamura et al., Adrenomedullin: a novel hypotensive peptide isolated from human pheochromocytoma, Biochem Biophys Res Commun
Koff, Schenkelberg, Williams, Baric, Mcdermott et al., Development and deployment of COVID-19 vaccines for those most vulnerable, Sci Transl Med
Koo, Saraswati, Margolis, Immunolocalization of Bex protein in the mouse brain and olfactory system, J Comp Neurol
Kuang, Yang, Zhang, Zhang, Wu, Schlafen 1 Inhibits the Proliferation and Tube Formation of Endothelial Progenitor Cells, PLoS One
Kuchar, Miśkiewicz, Nitsch-Osuch, Szenborn, Pathophysiology of Clinical Symptoms in Acute Viral Respiratory Tract Infections
Kupershmidt, Su, Grewal, Sundaresh, Halperin et al., Ontology-Based Meta-Analysis of Global Collections of High-Throughput Public Data, PLoS One
Kurki, Vanderlaan, Dolbeare, Gray, Tan, Expression of proliferating cell nuclear antigen (PCNA)/cyclin during the cell cycle, Experimental Cell Research
Lamb, Crawford, Peck, Modell, Blat et al., The Connectivity Map: using geneexpression signatures to connect small molecules, genes, and disease, Science
Langreth, Waldholz, New era of personalized medicine: targeting drugs for each unique genetic profile, Oncologist
Lee, Farber, Localization of the gene for the human MIG cytokine on chromosome 4q21 adjacent to INP10 reveals a chemokine "mini-cluster, Cytogenet Cell Genet
Lee, Park, Jeong, Ahn, Choi et al., Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19, Sci Immunol
Leinonen, Sugawara, Shumway, The Sequence Read Archive, Nucleic Acids Research
Li, Hu, Wu, Yao, Li, Molecular Characteristics, Functions, and Related Pathogenicity of MERS-CoV Proteins, Engineering
Li, Huang, Chen, Peng, Roop et al., Apoptotic Cells Activate the "Phoenix Rising" Pathway to Promote Wound Healing and Tissue Regeneration, Science Signaling
Liao, Liu, Yuan, Wen, Xu et al., Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19, Nature Medicine
Liao, Wang, Jaehnig, Shi, Zhang, WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs, Nucleic Acids Res
Liu, Zhou, Wang, Zhang, Li, The Schlafen family: complex roles in different cell types and virus replication, Cell Biol Int
Lu, Zhao, Li, Niu, Yang et al., Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding, The Lancet
Marima, Hull, Penny, Dlamini, Mitotic syndicates Aurora Kinase B (AURKB) and mitotic arrest deficient 2 like 2 (MAD2L2) in cohorts of DNA damage response (DDR) and tumorigenesis, Mutation Research/Reviews in Mutation Research
Martincorena, Roshan, Gerstung, Ellis, Loo et al., High burden and pervasive positive selection of somatic mutations in normal human skin, Adv Virus Res
Mavrommatis, Fish, Platanias, The schlafen family of proteins and their regulation by interferons, J Interferon Cytokine Res
Memon, Bass, Moser, Fuller, Appel et al., Down-regulation of liver and heart specific fatty acid binding proteins by endotoxin and cytokines in vivo, Biochim Biophys Acta
Metzemaekers, Vanheule, Janssens, Struyf, Proost, Overview of the Mechanisms that May Contribute to the Non-Redundant Activities of Interferon-Inducible CXC Chemokine Receptor 3 Ligands, Front Immunol
Mittra, Samant, Sharma, Raghuram, Saha et al., Cell-free chromatin from dying cancer cells integrate into genomes of bystander healthy cells to induce DNA damage and inflammation, Cell Death Discovery
Mokhtari, Hassani, Ghaffari, Ebrahimi, Yarahmadi et al., COVID-19 and multiorgan failure: A narrative review on potential mechanisms, Journal of Molecular Histology
Naderi, Liu, Bennett, BEX2 regulates mitochondrial apoptosis and G1 cell cycle in breast cancer, Int J Cancer
Naderi, Molecular functions of brain expressed X-linked 2 (BEX2) in malignancies, Exp Cell Res
Nalepa, Barnholtz-Sloan, Enzor, Dey, He et al., The tumor suppressor CDKN3 controls mitosis, J Cell Biol
Okamoto, Kitabayashi, Taya, KAP1 dictates p53 response induced by chemotherapeutic agents via Mdm2 interaction, Biochemical and Biophysical Research Communications
Pedregosa, Varoquaux, Gramfort, Michel, Thirion et al., Scikit-learn: Machine Learning in Python, JMLR
Perlman, Dandekar, Immunopathogenesis of coronavirus infections: implications for SARS, Nat Rev Immunol
Petersen, Koopmans, Go, Hamer, Petrosillo et al., Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics, Lancet Infect Dis
Phipps, Dubrana, DNA Repair in Space and Time: Safeguarding the Genome with the Cohesin Complex, Genes
Puck, Aigner, Modak, Cejka, Blaas et al., Expression and regulation of Schlafen (SLFN) family members in primary human monocytes, monocyte-derived dendritic cells and T cells, Results Immunol
Pulit-Penaloza, Scherbik, Brinton, Activation of Oas1a gene expression by type I IFN requires both STAT1 and STAT2 while only STAT2 is required for Oas1b activation, Virology
Radhakrishnan, Feliciano, Najmabadi, Haegebarth, Kandel et al., Constitutive expression of E2F-1 leads to p21-dependent cell cycle arrest in S phase of the cell cycle, Oncogene
Rahimmanesh, Shariati, Dana, Esmaeili, Vaseghi et al., Cancer Occurrence as the Upcoming Complications of COVID-19, Front Mol Biosci
Raj, Mou, Smits, Dekkers, Müller et al., Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC, Nature
Ramasamy, Mondry, Holmes, Altman, Key Issues in Conducting a Meta-Analysis of Gene Expression Microarray Datasets, PLoS Med
Roberts, Deming, Paddock, Cheng, Yount et al., A Mouse-Adapted SARS-Coronavirus Causes Disease and Mortality in BALB/c Mice, PLoS Pathog
Rockx, Baas, Zornetzer, Haagmans, Sheahan et al., Early Upregulation of Acute Respiratory Distress Syndrome-Associated Cytokines Promotes Lethal Disease in an Aged-Mouse Model of Severe Acute Respiratory Syndrome Coronavirus Infection, J Virol
Ryan, Fernandez, Peterson, Sheneman, Podell et al., Activation of S6 signaling is associated with cell survival and multinucleation in hyperplastic skin after epidermal loss of AURORA-A Kinase, Cell Death Differ
Saijoh, Nakamura-Shima, Shibuya-Takahashi, Ito, Sugawara et al., Discovery of a chemical compound that suppresses expression of BEX2, a dormant cancer stem cellrelated protein, Biochem Biophys Res Commun
Schindler, Darnell, Transcriptional responses to polypeptide ligands: the JAK-STAT pathway, Annu Rev Biochem
Schmid, Stijnen, White, Handbook of Meta-Analysis
Schmitz, Watrin, Lénárt, Mechtler, Peters, Sororin Is Required for Stable Binding of Cohesin to Chromatin and for Sister Chromatid Cohesion in Interphase, Current Biology
Schulte-Schrepping, Reusch, Paclik, Baßler, Schlickeiser et al., Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment, Cell
Schwarz, Katayama, Hedrick, Schlafen, a New Family of Growth Regulatory Genes that Affect Thymocyte Development, Immunity
Sehgal, Metastable biomolecular condensates of interferon-inducible antiviral Mx-family GTPases: A paradigm shift in the last three years, J Biosci
Shabrish, Mittra, Cytokine Storm as a Cellular Response to dsDNA Breaks: A New Proposal, Front Immunol
Shereen, Khan, Kazmi, Bashir, Siddique, COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses, J Adv Res
Sievert, Interactive web-based data visualization with R, plotly, and shiny
Singh, Singh, Sarma, Batra, Joshi et al., A Comprehensive Review of Animal Models for Coronaviruses: SARS-CoV-2, SARS-CoV, and MERS-CoV, Virol Sin
Song, Xu, Bao, Zhang, Yu et al., From SARS to MERS, Thrusting Coronaviruses into the Spotlight, Viruses
Sparks, Lau, Tsang, Expanding the Immunology Toolbox: Embracing Public-Data Reuse and Crowdsourcing, Immunity
Stoeber, Tlsty, Happerfield, Thomas, Romanov et al., DNA replication licensing and human cell proliferation, Journal of Cell Science
Subramaniyan, Larabee, Bodas, Moore, Burgett et al., Characterization of the SARS-CoV-2 Host Response in Primary Human Airway Epithelial Cells from Aged Individuals, Viruses
Sulli, Micco, Di, Fda, Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer, Nature Reviews Cancer
Szklarczyk, Gable, Lyon, Junge, Wyder et al., STRING v11: proteinprotein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets, Nucleic Acids Research
Takahashi, Ellingson, Wong, Israelow, Lucas et al., Sex differences in immune responses that underlie COVID-19 disease outcomes, Nature
Tan, Lim, Hong, Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus, Antiviral Res
Tokunaga, Zhang, Naseem, Puccini, Berger et al., CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation -A target for novel cancer therapy, Cancer Treat Rev
Totura, Bavari, Broad-spectrum coronavirus antiviral drug discovery, Expert Opin Drug Discov
Tsuruda, Kato, Kuwasako, Kitamura, Adrenomedullin: Continuing to explore cardioprotection, Peptides
Vazirinejad, Ahmadi, Arababadi, Hassanshahi, Kennedy, The biological functions, structure and sources of CXCL10 and its outstanding part in the pathophysiology of multiple sclerosis, Neuroimmunomodulation
Waga, Hannon, Beach, Stillman, The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA, Nature
Wan, Shang, Graham, Baric, Li, Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus, J Virol
Wang, Bonkovsky, De Lemos, Burczynski, Recent insights into the biological functions of liver fatty acid binding protein 1, J Lipid Res
Wang, Vasaikar, Shi, Greer, Zhang, WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit, Nucleic Acids Res
Weiss, Davies, Karwan, Ileva, Ozaki et al., Itaconic acid mediates crosstalk between macrophage metabolism and peritoneal tumors, J Clin Invest
Wilk, Rustagi, Zhao, Roque, Martínez-Colón et al., A single-cell atlas of the peripheral immune response in patients with severe COVID-19, Nature Medicine
Woods, Gladfelter, The state of the septin cytoskeleton from assembly to function, Current Opinion in Cell Biology
Wu, Chen, Wang, Tang, Kang, ACOD1 in immunometabolism and disease, Cell Mol Immunol
Wu, Peng, Huang, Ding, Wang et al., Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China, Cell Host Microbe
Yamamoto-Shimojima, Osawa, Saito, Yamamoto, Induced pluripotent stem cells established from a female patient with Xq22 deletion confirm that BEX2 escapes from X-chromosome inactivation, Congenit Anom
Zaim, Chong, Sankaranarayanan, Harky, COVID-19 and Multiorgan Response, Current Problems in Cardiology
Zaki, Van Boheemen, Bestebroer, Osterhaus, Fouchier, Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia, N Engl J Med
Zekavat, Lin, Bick, Liu, Paruchuri et al., Hematopoietic mosaic chromosomal alterations increase the risk for diverse types of infection, Nature Medicine
Zhou, Yang, Wang, Hu, Zhang et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature
Zornetzer, Frieman, Rosenzweig, Korth, Page et al., Transcriptomic analysis reveals a mechanism for a prefibrotic phenotype in STAT1 knockout mice during severe acute respiratory syndrome coronavirus infection, J Virol
DOI record:
{
"DOI": "10.1093/biomethods/bpad033",
"ISSN": [
"2396-8923"
],
"URL": "http://dx.doi.org/10.1093/biomethods/bpad033",
"abstract": "<jats:title>Abstract</jats:title>\n <jats:p>The emergence of SARS-CoV-2 reawakened the need to rapidly understand the molecular etiologies, pandemic potential, and prospective treatments of infectious agents. The lack of existing data on SARS-CoV-2 hampered early attempts to treat severe forms of COVID-19 during the pandemic. This study coupled existing transcriptomic data from SARS-CoV-1 lung infection animal studies with crowdsourcing statistical approaches to derive temporal meta-signatures of host responses during early viral accumulation and subsequent clearance stages. Unsupervised and supervised machine learning approaches identified top dysregulated genes and potential biomarkers (e.g., CXCL10, BEX2, and ADM). Temporal meta-signatures revealed distinct gene expression programs with biological implications to a series of host responses underlying sustained Cxcl10 expression and Stat signaling. Cell cycle switched from G1/G0 phase genes, early in infection, to a G2/M gene signature during late infection that correlated with the enrichment of DNA Damage Response and Repair genes. The SARS-CoV-1 meta-signatures were shown to closely emulate human SARS-CoV-2 host responses from emerging RNAseq, single cell and proteomics data with early monocyte-macrophage activation followed by lymphocyte proliferation. The circulatory hormone adrenomedullin was observed as maximally elevated in elderly patients that died from COVID-19. Stage-specific correlations to compounds with potential to treat COVID-19 and future coronavirus infections were in part validated by a subset of twenty-four that are in clinical trials to treat COVID-19. This study represents a roadmap to leverage existing data in the public domain to derive novel molecular and biological insights and potential treatments to emerging human pathogens.</jats:p>",
"author": [
{
"affiliation": [
{
"name": "Illumina Corporation , San Diego, CA"
}
],
"family": "Flynn",
"given": "James",
"sequence": "first"
},
{
"affiliation": [
{
"name": "University of Colorado Anschutz Medical Campus Gates Center for Regenerative Medicine, , Aurora, CO"
}
],
"family": "Ahmadi",
"given": "Mehdi M",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Bioinfo Solutions LLC , Parker, CO"
}
],
"family": "McFarland",
"given": "Chase T",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Illumina Corporation , San Diego, CA"
}
],
"family": "Kubal",
"given": "Michael D",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Bioinfo Solutions LLC , Parker, CO"
}
],
"family": "Taylor",
"given": "Mark A",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Illumina Corporation , San Diego, CA"
}
],
"family": "Cheng",
"given": "Zhang",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "University of Colorado Anschutz Medical Campus Gates Center for Regenerative Medicine, , Aurora, CO"
}
],
"family": "Torchia",
"given": "Enrique C",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Bioinfo Solutions LLC , Parker, CO"
}
],
"family": "Edwards",
"given": "Michael G",
"sequence": "additional"
}
],
"container-title": "Biology Methods and Protocols",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2023,
11,
14
]
],
"date-time": "2023-11-14T18:44:22Z",
"timestamp": 1699987462000
},
"deposited": {
"date-parts": [
[
2023,
11,
14
]
],
"date-time": "2023-11-14T18:44:22Z",
"timestamp": 1699987462000
},
"indexed": {
"date-parts": [
[
2023,
11,
15
]
],
"date-time": "2023-11-15T00:42:21Z",
"timestamp": 1700008941804
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2023,
11,
14
]
]
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "am",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
11,
14
]
],
"date-time": "2023-11-14T00:00:00Z",
"timestamp": 1699920000000
}
}
],
"link": [
{
"URL": "https://academic.oup.com/biomethods/advance-article-pdf/doi/10.1093/biomethods/bpad033/53403496/bpad033.pdf",
"content-type": "application/pdf",
"content-version": "am",
"intended-application": "syndication"
},
{
"URL": "https://academic.oup.com/biomethods/advance-article-pdf/doi/10.1093/biomethods/bpad033/53403496/bpad033.pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "286",
"original-title": [],
"prefix": "10.1093",
"published": {
"date-parts": [
[
2023,
11,
14
]
]
},
"published-online": {
"date-parts": [
[
2023,
11,
14
]
]
},
"publisher": "Oxford University Press (OUP)",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://academic.oup.com/biomethods/advance-article/doi/10.1093/biomethods/bpad033/7420215"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"General Agricultural and Biological Sciences",
"General Biochemistry, Genetics and Molecular Biology"
],
"subtitle": [],
"title": "Crowdsourcing Temporal Transcriptomic Coronavirus Host Infection Data: resources, guide, and novel insights",
"type": "journal-article"
}